Accurate models of the cross talk between signaling pathways and transcriptional regulatory networks within cells are essential to understand complex response programs. We present a new computational method that combines condition-specific time series expression data with general protein interaction data to reconstruct dynamic and causal stress response networks. These networks characterize the pathways involved in the response, their time of activation, and the affected genes. The signaling and regulatory components of our networks are linked via a set of common transcription factors that serve as targets in the signaling network and as regulators of the transcriptional response network. SDREM has been applied to infer the pathways involved in yeast stress responses and the human immune response to viral infection.
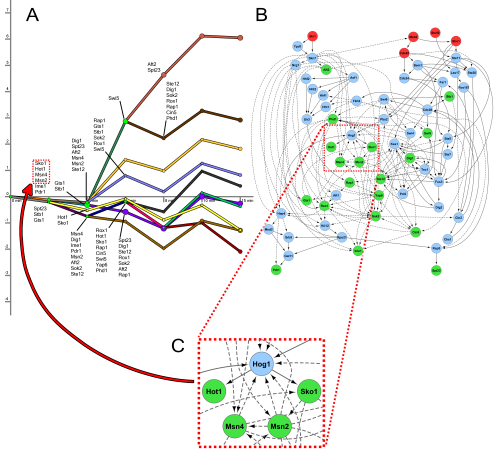
The figure above shows the regulatory paths (left) and signaling network (right) that SDREM learns in the yeast osmotic stress response.
SDREM has been described in
- Linking the signaling cascades and dynamic regulatory networks controlling stress responses.
Anthony Gitter, Miri Carmi, Naama Barkai, Ziv Bar-Joseph.
Genome Research. 23:2, 2013.
- Identifying proteins controlling key disease signaling pathways.
Anthony Gitter, Ziv Bar-Joseph.
Bioinformatics. 29:13, 2013.
21st Annual International Conference on Intelligent Systems for Molecular Biology/12th European Conference on Computational Biology (ISMB/ECCB 2013) Proceedings.
- The SDREM method for reconstructing signaling and regulatory response networks: Applications for studying disease progression.
Anthony Gitter, Ziv Bar-Joseph.
In Systems Biology of Alzheimer's Disease, Methods in Molecular Biology series. Vol 1303, pp 493-506, 2015.
MT-SDREM is a SDREM extension that jointly models multiple conditions using a multi-task learning framework. It can be downloaded at the MT-SDREM site and is described in