Dynamic Regulatory Events Miner (DREM)
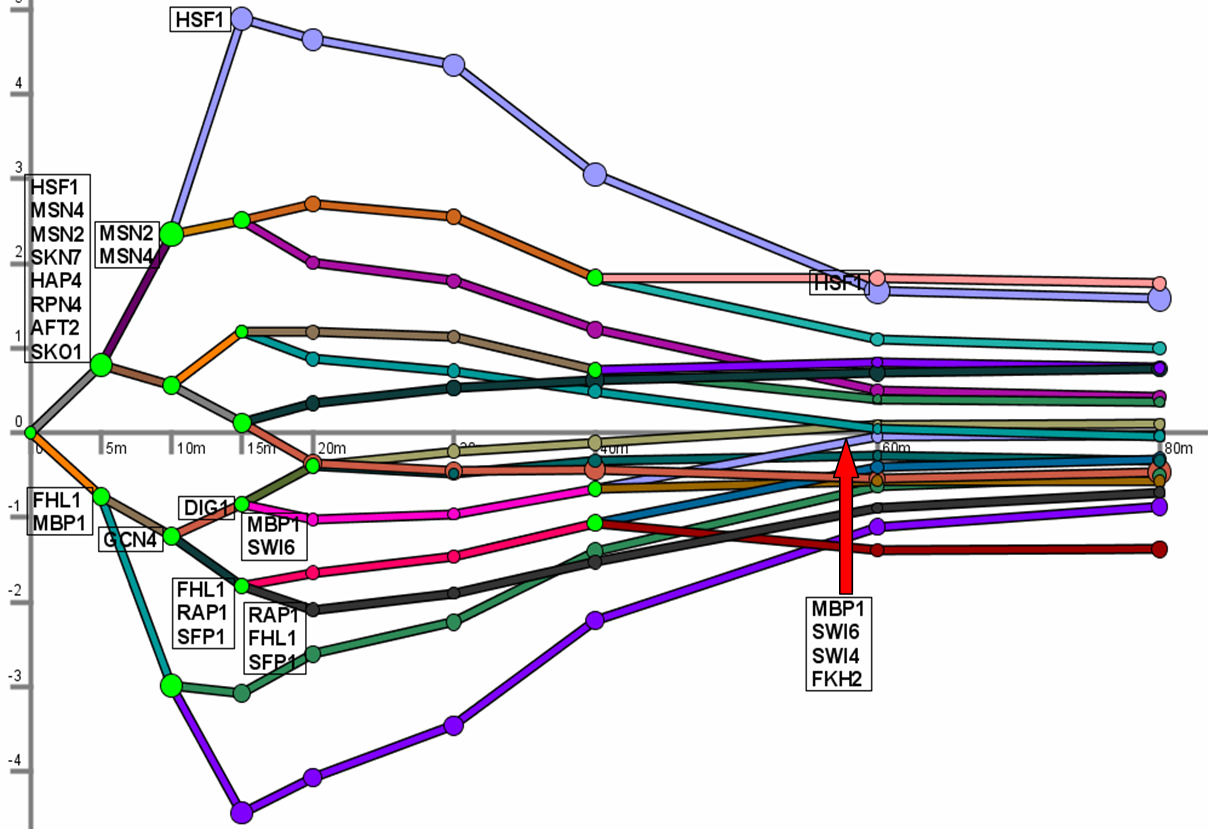
The Dynamic Regulatory Events Miner (DREM) allows one to model, analyze,
and visualize
transcriptional gene regulation dynamics. The method of DREM takes as input time series gene expression
data and static or dynamic transcription factor-gene interaction data (e.g. ChIP-seq data), and
produces as output a dynamic regulatory map.
The dynamic regulatory map highlights major bifurcation events in the time series expression data
and transcription factors potentially responsible for them. See the manual and papers below for more
details.
DREM 2.0 was released and supports a number of new features including (see manual for details):
- new static binding data for mouse, human, D. melanogaster, A. thaliana
- a new and more flexible implementation of the IOHMM supports dynamic binding data for each time point or as a mix of static/dynamic TF input
- expression levels of TFs can be used to improve the models learned by DREM
- the motif finder DECOD can be used in conjunction with DREM and help find DNA motifs for unannotated splits
- new features for the visualization of expressed TFs, dragging boxes in the model view, and switching between representations
Click here to download DREM version 2.0.4
(versionlog.txt)
- DREM User manual: DREMmanual.pdf
-
The DREM method was originally described and applied to yeast in the paper:
J. Ernst, O. Vainas, C.T. Harbison, I. Simon, and Z. Bar-Joseph.
Reconstructing Dynamic Regulatory Maps.
Nature-EMBO Molecular Systems Biology, 3:74, 2007. Click
here for Supporting Information
specific to that paper
- If you use the DREM 2.0 and beyond please cite also:
M.H. Schulz, W.E. Devanny, A. Gitter, S. Zhong, J. Ernst and Z. Bar-Joseph
DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data,
BMC Systems Biology 2012
- Other examples of published papers using DREM as part of the data analysis can be found
here.
-
Sign-up here for a google group mailing list to receive announcements of new versions of DREM and also STEM.
- Email any questions, comments, or bugs found to
Jason Ernst
(jernst@cs.cmu.edu) or Marcel Schulz (marcel.schulz@em.uni-frankfurt.de)
- Source code is available on GitHub here.
- DREM is now available under a GPL v3.0 license
Short Primer: Starting DREM
1. To run DREM Java 1.5 or higher must be installed
2. Save the drem2.zip file to a computer.
3. Unzip the drem2.zip file
4a. If on windows double click on the .cmd file
corresponding to the condition of interest to see
DREM opened with the data from that condition.
4b. If not on windows start DREM from the drem directory with the command
java -mx1024M -ms512M -jar drem.jar -d [defaultfilename]
where defaultfilename is the name of the condition DREM should be
opened with. For instance to open DREM with the heat shock data type
java -mx1024M -ms512M -jar drem.jar -d defaultsHeatSample.txt
If you want to use a human dataset try java -mx1024M -ms512M -jar drem.jar -d defaultsHumanSample.txt
Using DREM
5. Once DREM is open press the execute button at the bottom.
6. To see only genes assigned to a certain path click on that
path.
7. The gene table button displays the names of the genes currently
display by DREM.
8. The GO table displays a GO enrichment for all genes displayed by
DREM
Please read the provided manual to learn more about the features of DREM!
DREM extensions
DREM has been extended to model additional biological settings and other types of high-throughput data. Details, code, and references can be found on the respective web pages:
- mirDREM: supports the use of microRNAs in DREM allowing for dynamic models that integrate both TFs and miRNAs
- SDREM: links dynamic transcriptional regulatory networks to upstream signaling pathways
- MT-SDREM: multitask learning for modeling multiple conditions with SDREM
- SMARTS: reconstructs and clusters multiple condition-specific transcriptional response networks
- cDREM: models combinatorial gene regulation
- iDREM: interactive visualization and integration of protein interaction and epigenetic data with DREM
The development of DREM, DREM2.0 and mirDREM was supported in part by NIH grants NO1 AI-5001, 1R01GM085022 and U01HL108642 and NSF CAREER award 0448453 to ZBJ
