 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| DME9: | --AWGAAAAA-- |
| DME14: | ---AGRAGCTG- |
| DME23: | --AAGANGAA-- |
| DME29: | -AGAGAAAR--- |
| DME80: | GAMAGCAG---- |
| DME38: | -TGGGAAAR--- |
| DME54: | CTCACATW---- |
| DME64: | GGAAGAAG---- |
| DME61: | --CARCAGCA-- |
| DME2: | ----GAAAAAWG |
| Familial Profile: (click for matrix) | 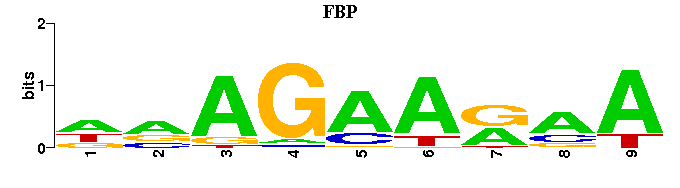 |
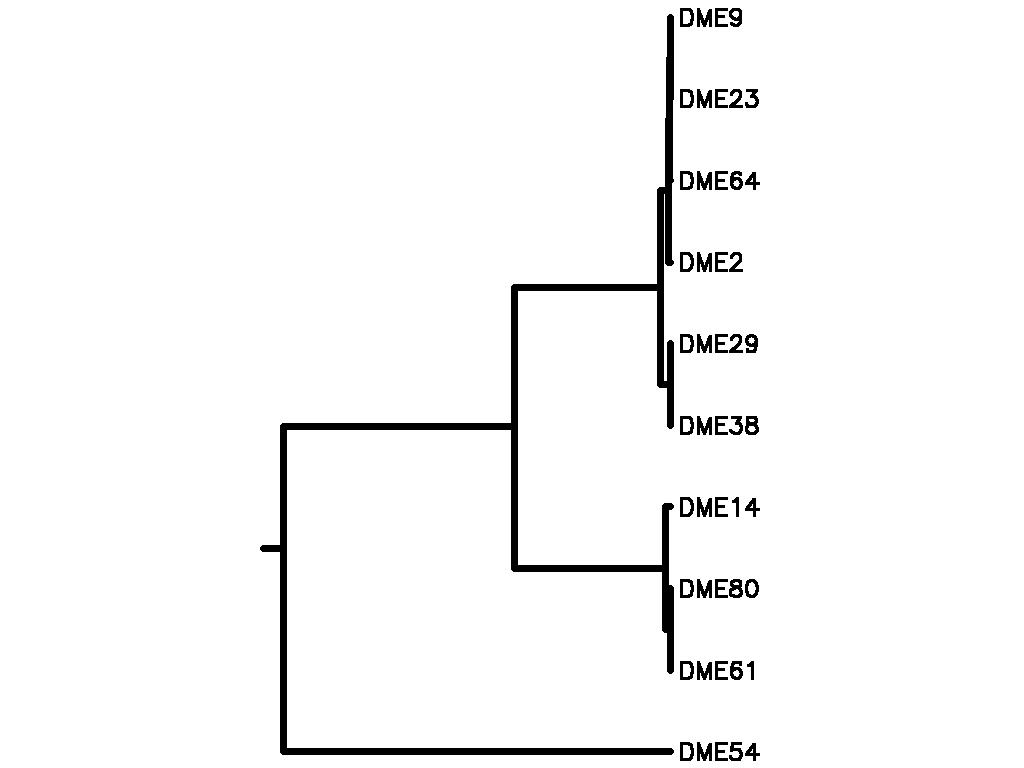
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| DME9 | 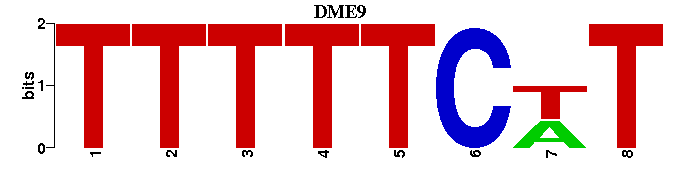 | 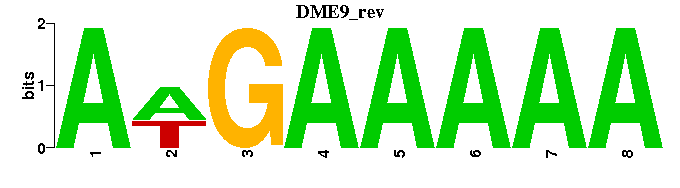 |
| Name | E value | |||
| Helios_M01004 | 1.3019e-06 | NTTTTCCTWN | 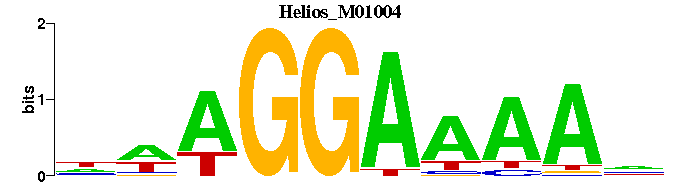 | |
| Rim101p_M01030 | 1.4515e-05 | --TTNCTTG | 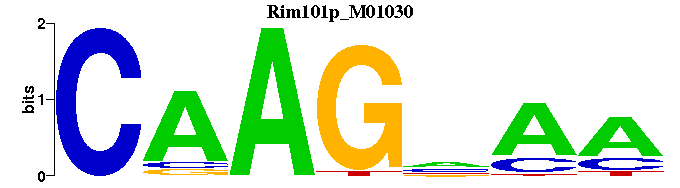 | |
| dl_M00043 | 1.7481e-05 | NGGAAAAACC | 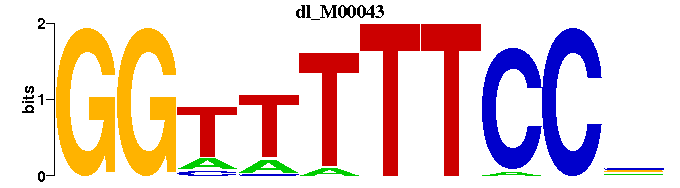 | |
| AG_M01133 | 8.1139e-05 | TWNCAAAAAWGGNAAN | 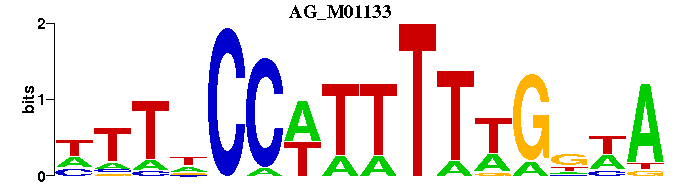 | |
| Evi-1_M00011 | 1.0421e-04 | -TTATCTTG |  |
| DME23 | 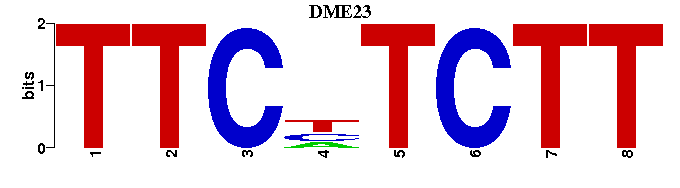 | 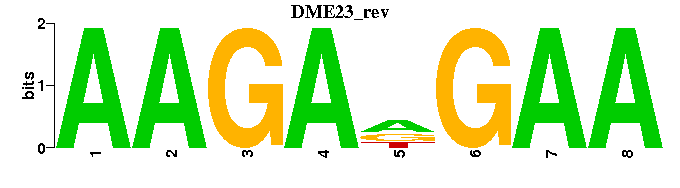 |
| Name | E value | |||
| Elf-1_M00746 | 3.8950e-08 | AYTTCCTCTTN |  | |
| PEND_M01015 | 2.0893e-06 | AAGAAGT- |  | |
| XPF-1_M00684 | 2.3251e-05 | GKTSNYCNG |  | |
| PU.1_M00658 | 4.0255e-05 | CTTCCTC-- |  | |
| dTCF_M00362 | 1.0869e-04 | AAGATCAAAG |  |
| DME64 | 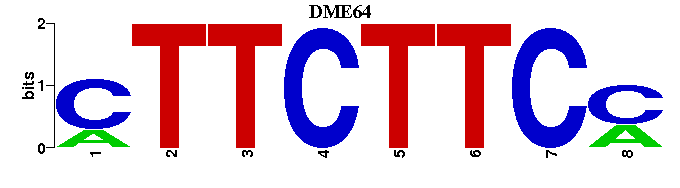 | 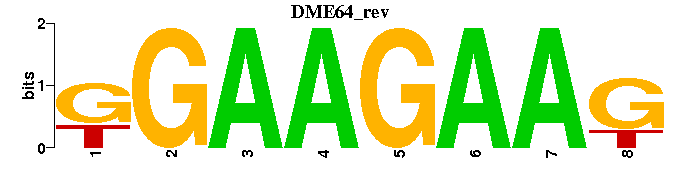 |
| Name | E value | |||
| PEND_M01015 | 6.7563e-07 | --AAGAAGT |  | |
| PU.1_M00658 | 4.4447e-06 | -GAGGAAG |  | |
| Nrf-2_M00108 | 3.3753e-05 | -SNCTTCCGG |  | |
| Tel-2_M00678 | 1.3516e-04 | CAGGAAGTA- |  | |
| GCR1_M00046 | 3.2595e-04 | -GWGGAAGC | 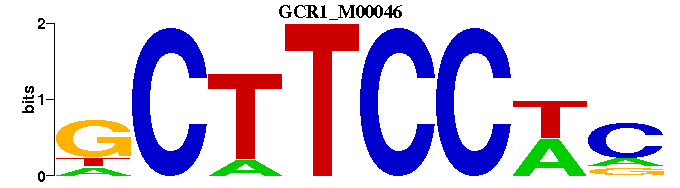 |
| DME2 | 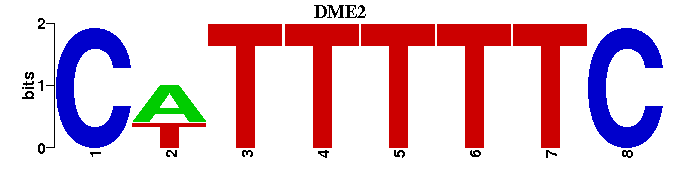 |  |
| Name | E value | |||
| MCM1+SFF_M01051 | 4.5626e-08 | TTRTTTANNTWTTNCCNNWTTNGGNA |  | |
| MADS-A_M00408 | 1.4529e-06 | TTTCCATTTTTNNWN |  | |
| AG_M01133 | 1.4630e-06 | TWNCAAAAAWGGNAAN |  | |
| MADS-B_M00404 | 2.0101e-06 | TTTSCATTTTTRGN |  | |
| dl_M00043 | 1.7481e-05 | GGTTTTTCCN |  |
| DME29 | 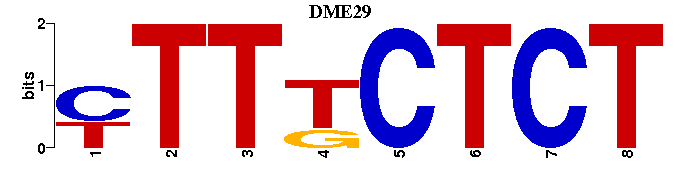 | 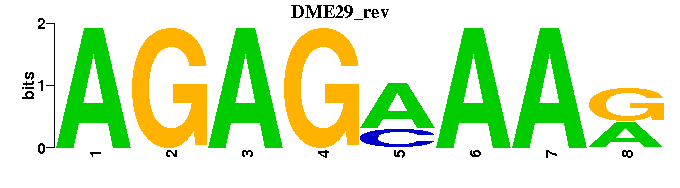 |
| Name | E value | |||
| Elf-1_M00746 | 1.9017e-06 | AYTTCCTCTTN |  | |
| BPC1_M01126 | 5.4962e-06 | NTTTC--- |  | |
| dl_M00120 | 5.3535e-05 | NGNTTTTCYC- |  | |
| IRF_M00972 | 6.0236e-05 | NTTTCWNTTT |  | |
| GAGA_M00723 | 6.0958e-05 | SWGAGMGNRA |  |
| DME38 | 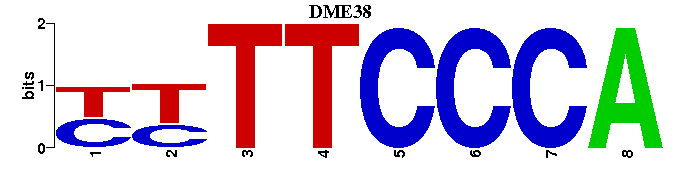 | 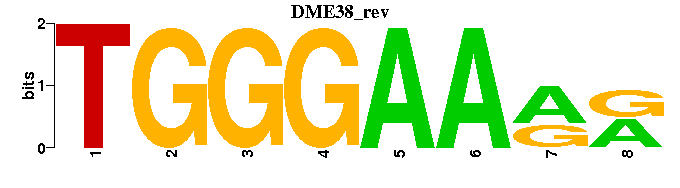 |
| Name | E value | |||
| Lyf-1_M00141 | 1.4081e-06 | -YCTCCCAA |  | |
| BPC1_M01126 | 4.3072e-06 | NTTTC--- |  | |
| RBP-Jkappa_M01112 | 9.2263e-06 | -KTTCCCACGN |  | |
| STAT1_M00492 | 1.7823e-05 | -SGGAANT |  | |
| ISGF-3_M00258 | 2.0473e-05 | -GNGAAANWGAAACT | 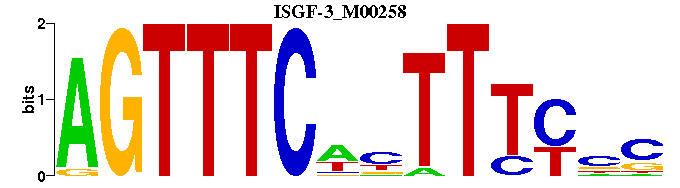 |
| DME14 | 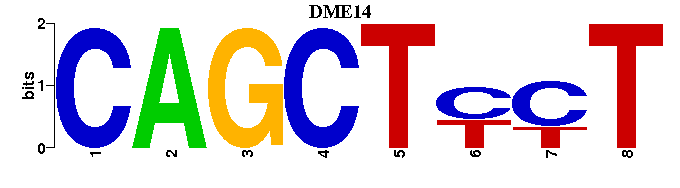 | 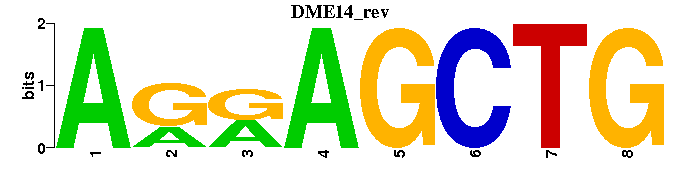 |
| Name | E value | |||
| LUN-1_M00480 | 2.7524e-05 | TCCCAAAGTAGCTGGG | 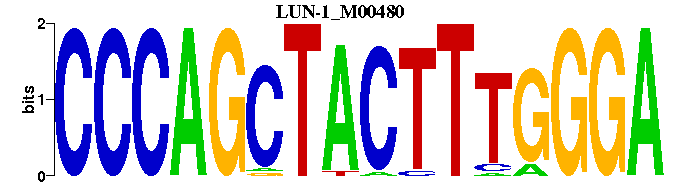 | |
| LBP-1_M00644 | 3.6468e-05 | SCAGCN--- |  | |
| Barbie_M00238 | 3.6633e-05 | CMNNCNSCTTTNNN | 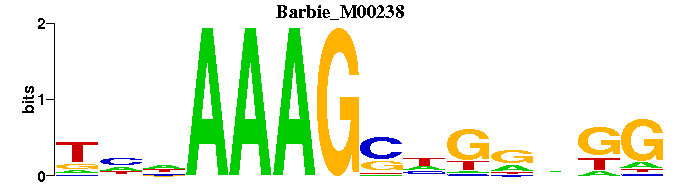 | |
| TAL1_M00993 | 5.9146e-05 | RNCAGNTGG |  | |
| AP-4_M00927 | 1.0269e-04 | -RCAGCTGN |  |
| DME80 | 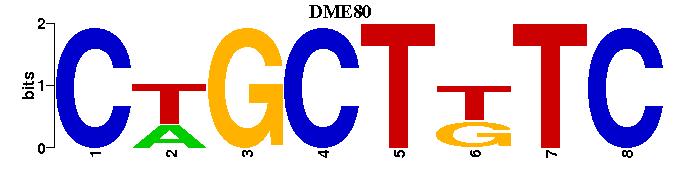 | 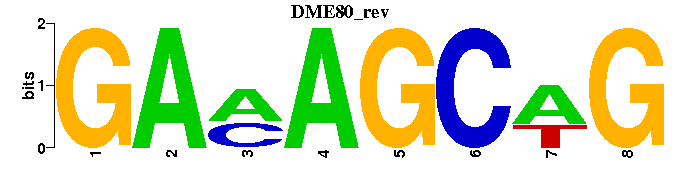 |
| Name | E value | |||
| LMAF_M01139 | 1.7859e-06 | STCAGCAG |  | |
| Barbie_M00238 | 2.8352e-05 | CMNNCNSCTTTNNN | 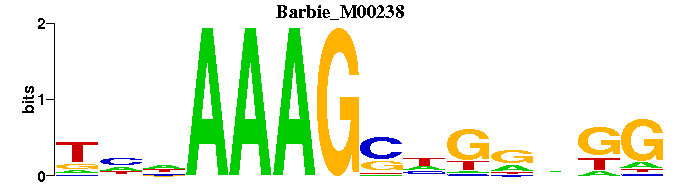 | |
| HEB_M00698 | 2.9896e-05 | --CAGCWGG |  | |
| BPC1_M01126 | 1.3462e-04 | ---NTTTC |  | |
| Grainyhead-Elf-1-NTF-1_M00951 | 1.3798e-04 | -AAACCRG |  |
| DME61 | 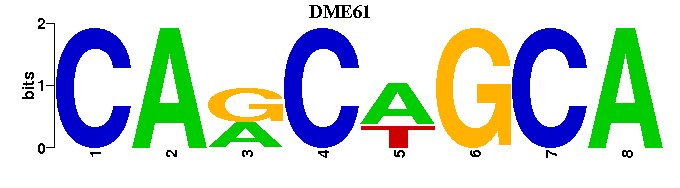 | 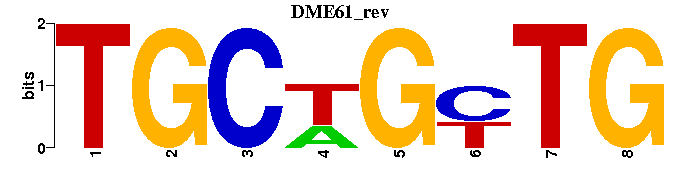 |
| Name | E value | |||
| HEB_M00698 | 5.7586e-06 | -CCWGCTG |  | |
| AP-4_M00927 | 8.8817e-06 | -RCAGCTGN |  | |
| myogenin_M00712 | 1.6420e-05 | CASCTGN- |  | |
| LMAF_M01139 | 3.7144e-05 | --CTGCTGAS |  | |
| NRSF_M01028 | 1.2170e-04 | YNCTGTCCRYGGTGCTGA | 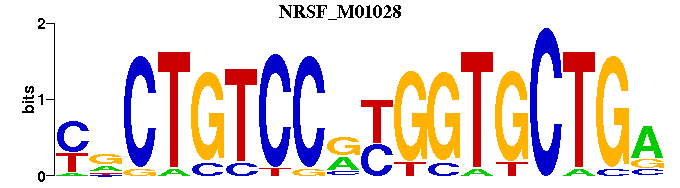 |
| DME54 |  |  |
| Name | E value | |||
| MIG1_M00061 | 4.5981e-05 | NNWWAAANSYGGGGNN |  | |
| CRP_M00202 | 9.9093e-05 | WANTGTGANNYNNNTCACAWTWATT | 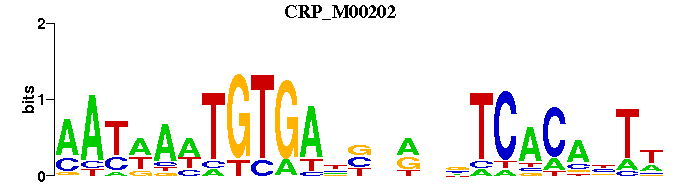 | |
| TFE_M01029 | 3.5993e-04 | -NCACATG |  | |
| IRF-1_M00747 | 4.0444e-04 | -AANTGA- |  | |
| IRF_M00972 | 6.0513e-04 | NTTTCWNTTT |  |