 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| DME10: | --CTTCAYTT--- |
| DME29: | ---TTTCCTKR-- |
| DME28: | -----TCCYTCWC |
| DME30: | ---TTTCCTGM-- |
| DME32: | CTTTTTGT----- |
| DME2: | ---TCTCTTTT-- |
| DME20: | -TTTTTGTT---- |
| DME75: | ---ATTCYTGT-- |
| DME5: | --TTTTGTTT--- |
| DME43: | --TTCTGGTK--- |
| Familial Profile: (click for matrix) | 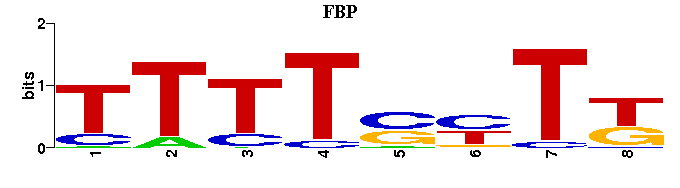 |
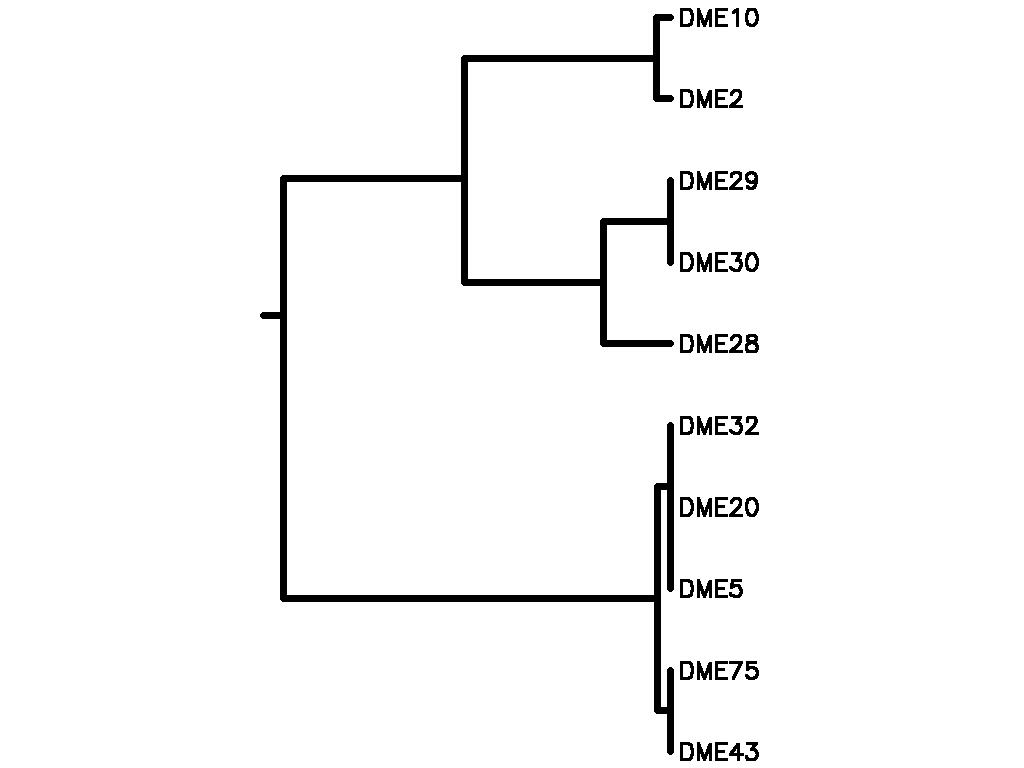
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| DME10 | 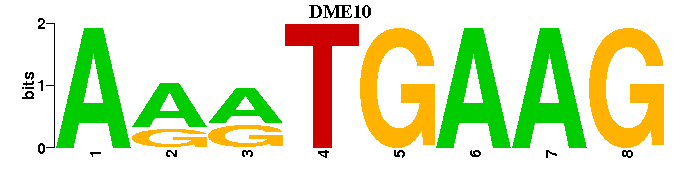 | 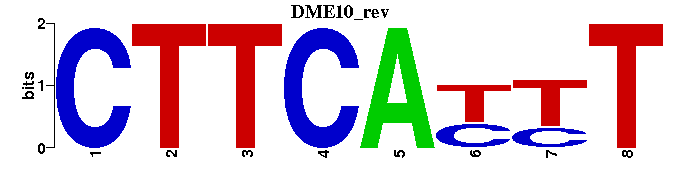 |
| Name | E value | |||
| ICSBP_M00699 | 1.4479e-07 | CAGTTTCAYTT | 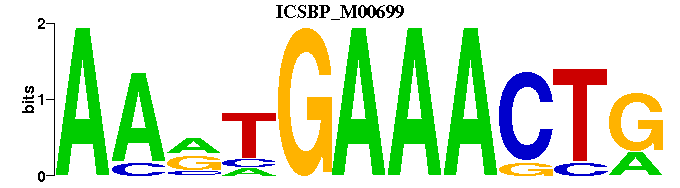 | |
| IRF_M00772 | 1.3014e-06 | NNNSTTTCANTTYY | 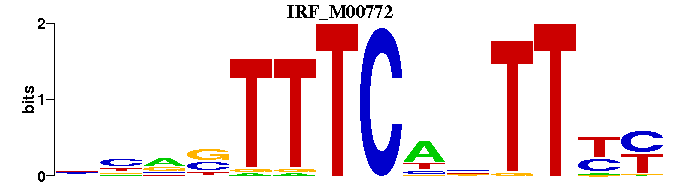 | |
| IRF-1_M00747 | 2.5909e-06 | AANTGA-- |  | |
| IRF_M00972 | 4.9058e-06 | NTTTCWNTTT | 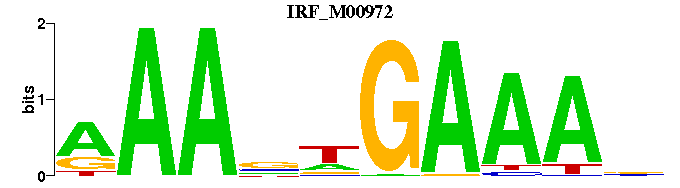 | |
| ISGF-3_M00258 | 1.2869e-05 | GNGAAANWGAAACT | 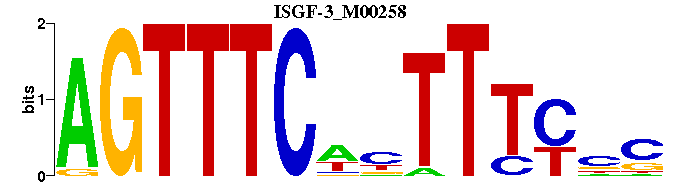 |
| DME2 | 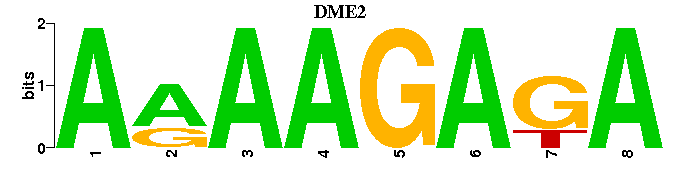 | 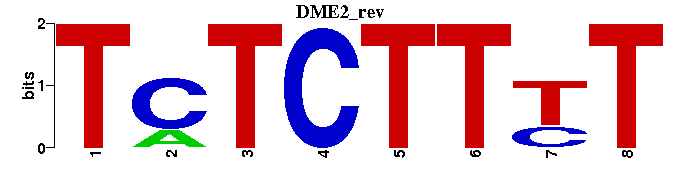 |
| Name | E value | |||
| ISGF-3_M00258 | 1.2221e-05 | GNGAAANWGAAACT |  | |
| IRF-2_M00063 | 1.2270e-05 | RSTTTCRCTTTT | 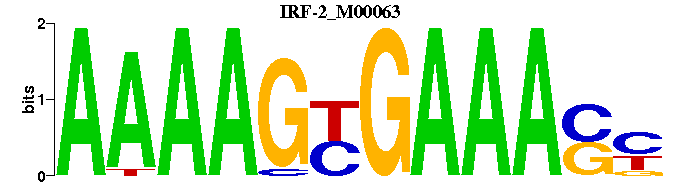 | |
| IRF-1_M00062 | 1.4614e-05 | GGTTTCRCTTTT |  | |
| GATA-4_M00632 | 2.9091e-05 | TSYCTKNTATC |  | |
| MEF-2_M00233 | 5.4562e-05 | NNNTTCTATTTTTAGTAACAN | 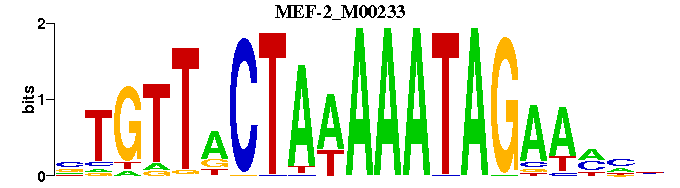 |
| DME29 | 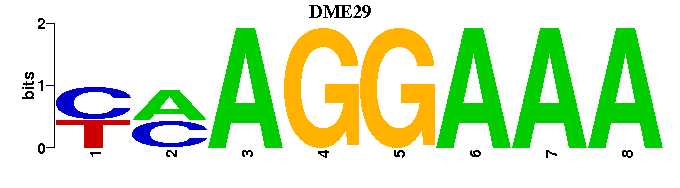 | 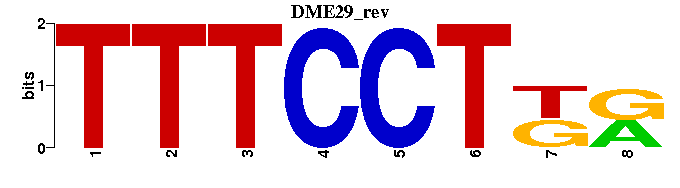 |
| Name | E value | |||
| STAT5B_M00459 | 2.5076e-06 | NWTTCCNGGAANYN | 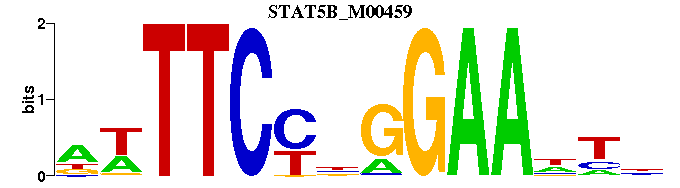 | |
| Helios_M01004 | 4.7483e-06 | NTTTTCCTWN | 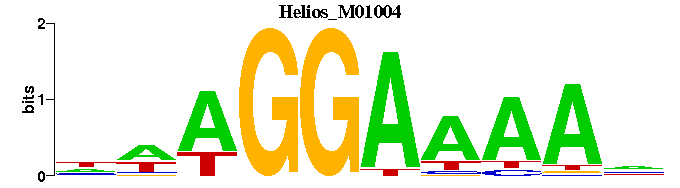 | |
| STE11_M01005 | 3.6230e-05 | TTTCTTTGTTC |  | |
| Tel-2_M00678 | 1.2298e-04 | -CAGGAAGTA |  | |
| STAT5A_M00457 | 1.2357e-04 | NRNTTCCNRGAANY | 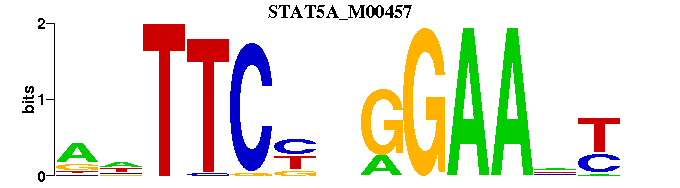 |
| DME30 | 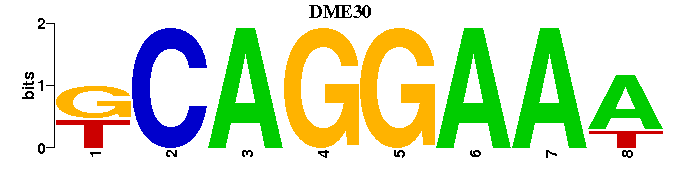 | 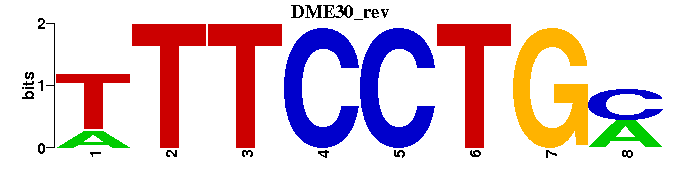 |
| Name | E value | |||
| STAT5B_M00459 | 1.0433e-06 | NRNTTCCNGGAAWN |  | |
| Tel-2_M00678 | 5.9898e-06 | -CAGGAAGTA |  | |
| Ttk_M00009 | 7.3629e-06 | GCAGGAC- |  | |
| STAT5A_M00457 | 2.0423e-05 | RNTTCYNGGAANYN |  | |
| Rim101p_M01030 | 2.5228e-05 | -CAAGNAA | 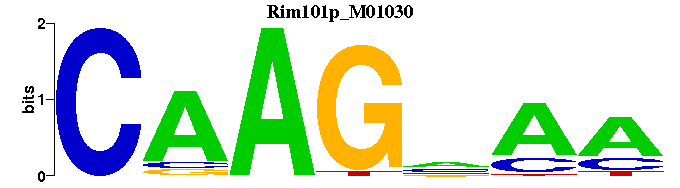 |
| DME28 | 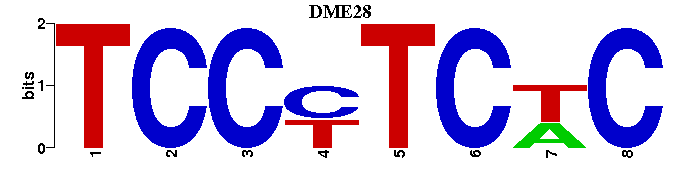 | 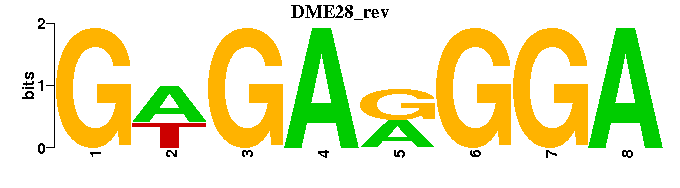 |
| Name | E value | |||
| MAF_M00648 | 6.3648e-05 | NSNMMACTTCCYYCN- | 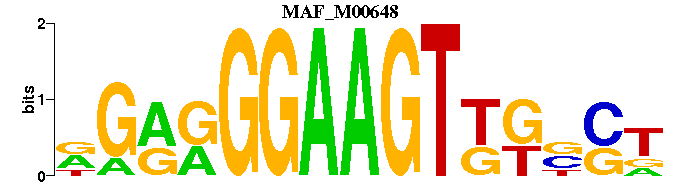 | |
| MAZ_M00649 | 1.1443e-04 | -CCCTCCC |  | |
| MZF1_M00084 | 3.0626e-04 | TTNCCCCTCNNN | 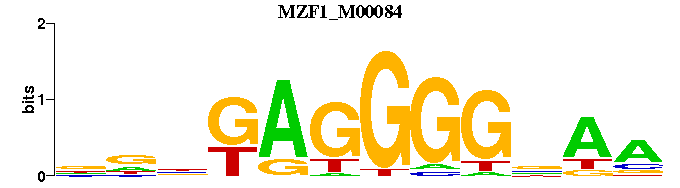 | |
| TFII-I_M00706 | 5.2599e-04 | CCTMCNTC-- |  | |
| Lmo2_M00278 | 5.7947e-04 | -CSNTATCK |  |
| DME32 |  | 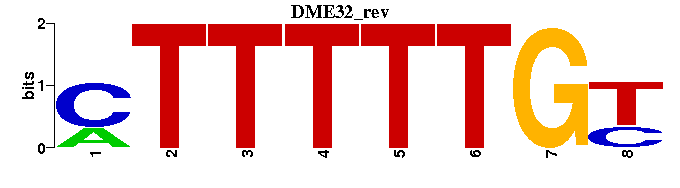 |
| Name | E value | |||
| Xvent-1_M00445 | 6.7077e-06 | RSRCAAATAGNN |  | |
| Hb_M00022 | 3.8260e-05 | NTTTTTNYK | 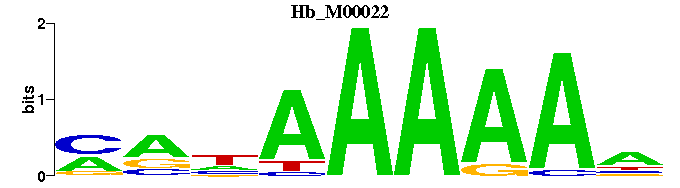 | |
| Dof_M01136 | 4.3812e-05 | NNNCTTTTTN- |  | |
| XFD-3_M00269 | 5.3609e-05 | -WKWTTGTTTACWN | 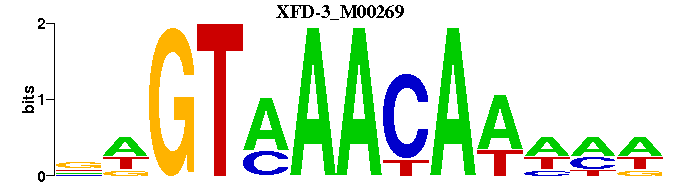 | |
| MADS-A_M00408 | 2.5286e-04 | TTTCCATTTTTNNWN |  |
| DME20 |  | 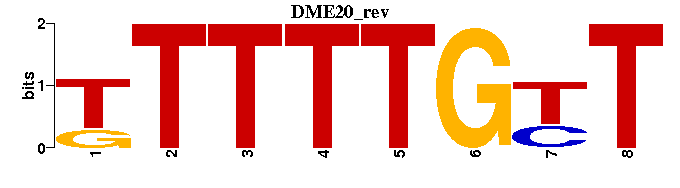 |
| Name | E value | |||
| XFD-3_M00269 | 5.7136e-07 | WKWTTGTTTACWN |  | |
| STE11_M01005 | 1.8232e-06 | GAACAAAGAAA |  | |
| SRY_M00148 | 2.3427e-06 | AACANM-- |  | |
| SOX_M01014 | 4.1764e-06 | NCNTAACAAAGA |  | |
| Hb_M00022 | 5.6917e-06 | NTTTTTNYK |  |
| DME5 |  | 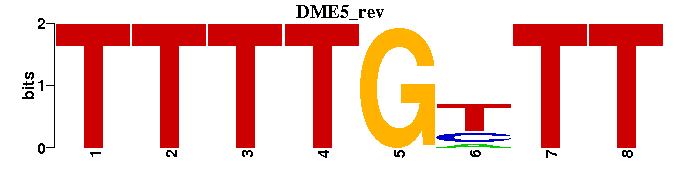 |
| Name | E value | |||
| BR-C_M00094 | 4.3946e-09 | TTWTSTTTAYWW |  | |
| XFD-3_M00269 | 1.5009e-08 | WKWTTGTTTACWN |  | |
| HFH4_M00742 | 7.3032e-08 | TAAACAAACAMW |  | |
| HNF3alpha_M00724 | 2.2382e-07 | NWRARCAAAY |  | |
| SRY_M00148 | 6.2508e-07 | -AACANM- |  |
| DME75 | 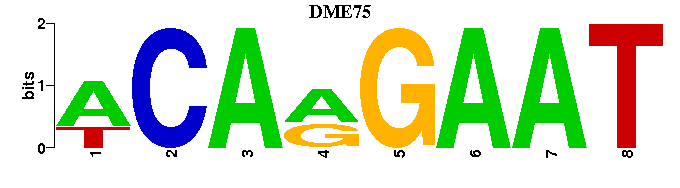 | 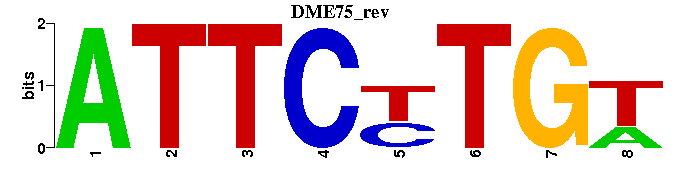 |
| Name | E value | |||
| Rim101p_M01030 | 7.2018e-06 | -CAAGNAA |  | |
| STAT5B_M00459 | 7.6219e-05 | NRNTTCCNGGAAWN |  | |
| ATHB-5_M00503 | 1.2227e-04 | -CAATNATT |  | |
| c-Ets-2_M00340 | 1.3601e-04 | RCAGGAAGTNNNT | 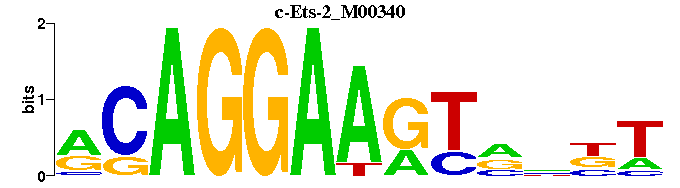 | |
| STAT5A_M00457 | 2.0686e-04 | NRNTTCCNRGAANY |  |
| DME43 |  | 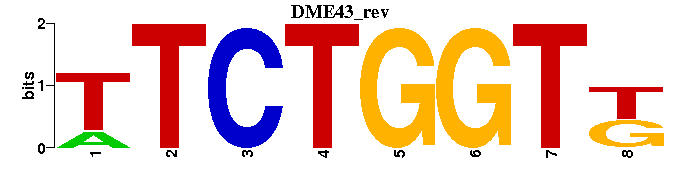 |
| Name | E value | |||
| Grainyhead-Elf-1-NTF-1_M00951 | 7.1404e-06 | --CYGGTTT |  | |
| Elk-1_M00007 | 2.7891e-04 | NNGNACTTCCKGTNN | 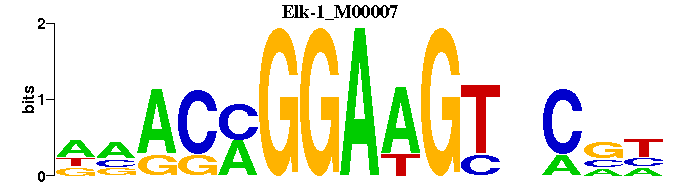 | |
| AML_M00769 | 5.0093e-04 | NNWRACCACANNNN |  | |
| STAT_M00777 | 5.5499e-04 | NNNNTTCTNGGA | 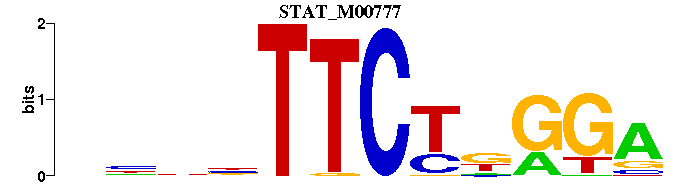 | |
| PEBP_M00984 | 8.6431e-04 | NTNACCACARNNNN |  |