 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| Motif1: | -GACATGTC---- |
| Motif2: | -----TGTGYCTG |
| Motif3: | -WAAATGTT---- |
| Motif4: | --CCCTGCAS--- |
| Motif5: | -GGCCWGGR---- |
| Motif6: | ----ATTTCWGA- |
| Motif7: | TCAGCTGA----- |
| Motif8: | --CTCTGTNT--- |
| Motif9: | ---TATTTGAA-- |
| Motif10: | -----TGTGGAAT |
| Familial Profile: (click for matrix) | 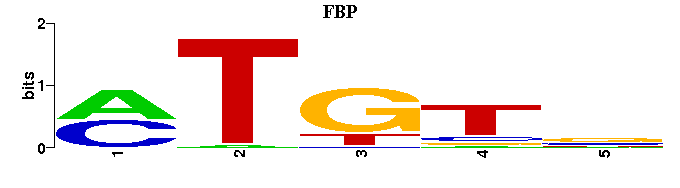 |
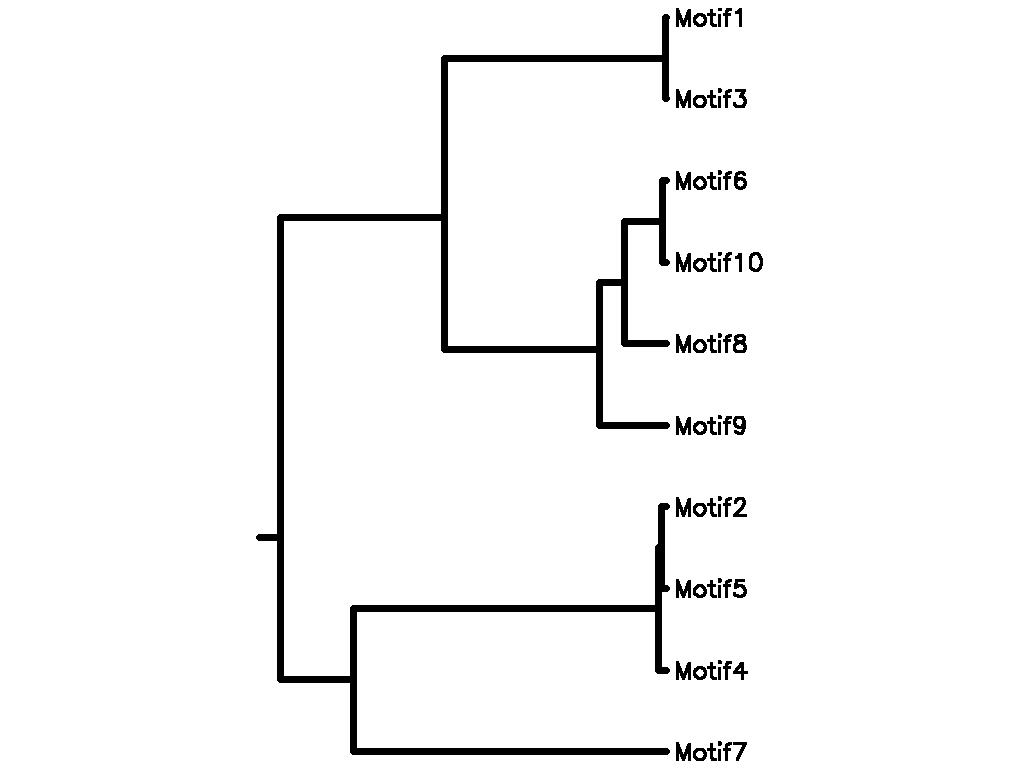
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| Motif1 | 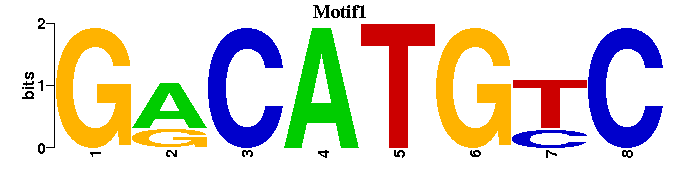 | 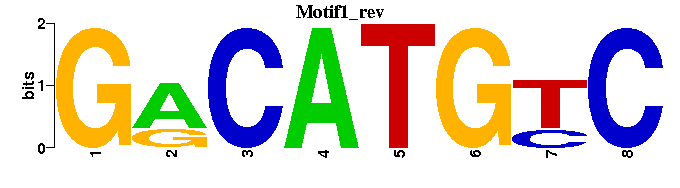 |
| Name | E value | |||
| p53_M00272 | 1.0929e-11 | GRCWTGYCY |  | |
| p53_M00761 | 7.3405e-10 | NGNCWTGYC | 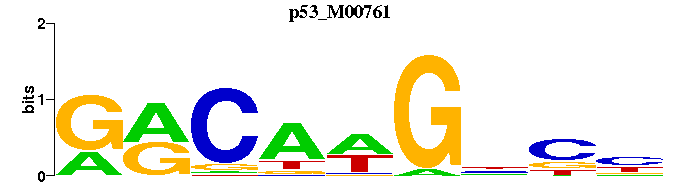 | |
| p53_M00034 | 5.7808e-08 | GACATGCCCGGGCATGTCY | 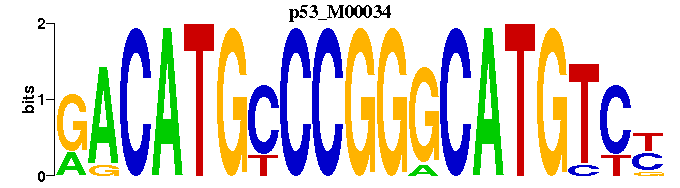 | |
| E2A_M00973 | 2.2670e-05 | RNCAGNT- |  | |
| XBP-1_M00251 | 1.0776e-04 | NWNNNMCACGTCANCN |  |
| Motif3 | 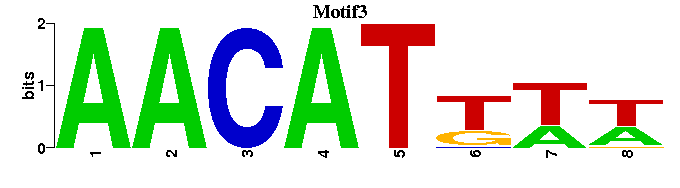 | 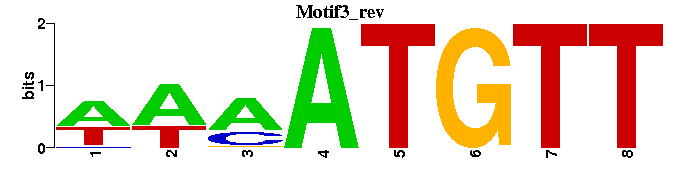 |
| Name | E value | |||
| AR_M00447 | 1.6343e-06 | AGAACAWNANGTWC | 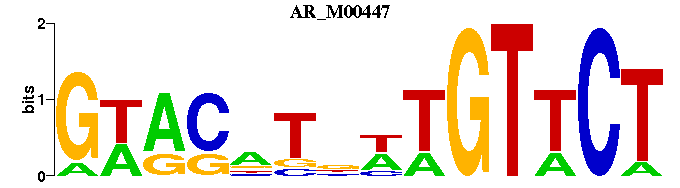 | |
| HNF3beta_M00131 | 9.1772e-06 | GNANTRTTTRYTTW | 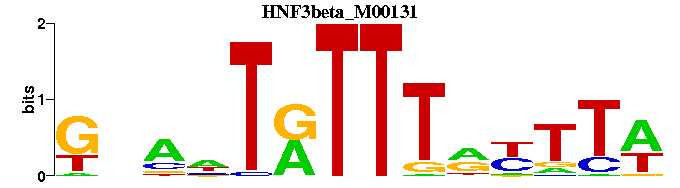 | |
| FOXP3_M00992 | 2.6294e-05 | NTMTGNNANAACNNNW- | 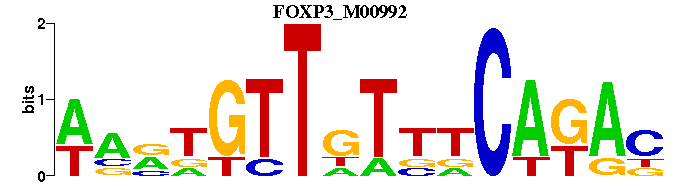 | |
| FOXJ2_M00423 | 4.5532e-05 | NMAAATATTATKR | 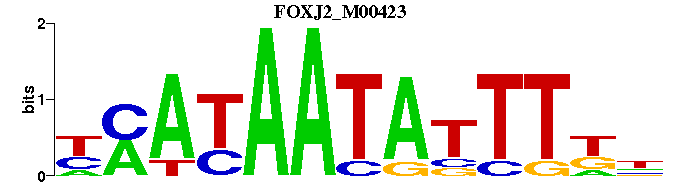 | |
| DMRT3_M01148 | 1.4715e-04 | NWTTGWTACATTNN | 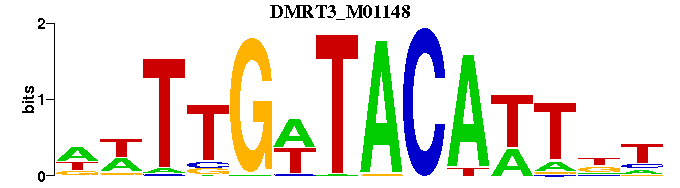 |
| Motif6 | 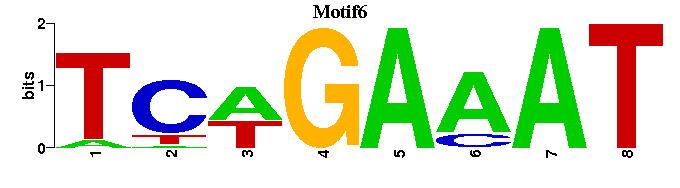 | 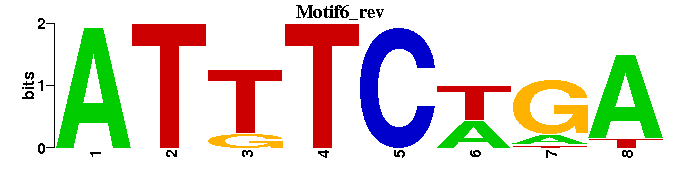 |
| Name | E value | |||
| C-EBPgamma_M00622 | 5.0943e-08 | WTTYTGAAATNA | 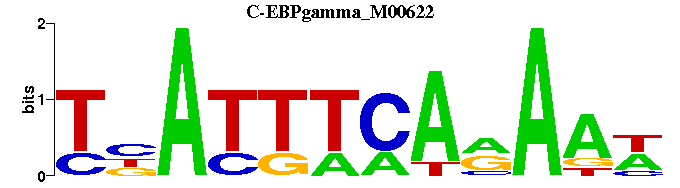 | |
| FOXP3_M00992 | 5.1979e-05 | WNNNGTTNTNNCAKAN |  | |
| TCF11-MafG_M00284 | 1.2386e-04 | CNNANNTGCTGNGTCATNNNN | 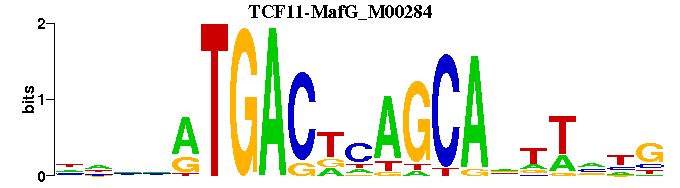 | |
| Pax-5_M00144 | 2.3485e-04 | NNNRYNGTYMCGCTYNANNNNNCNNNY | 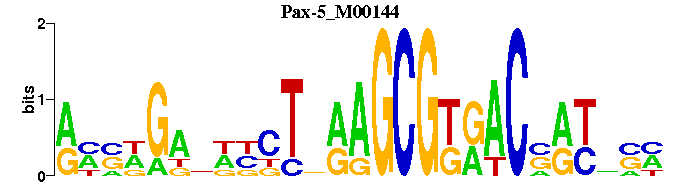 | |
| HMG_M00750 | 3.4306e-04 | AWTTTC--- |  |
| Motif10 | 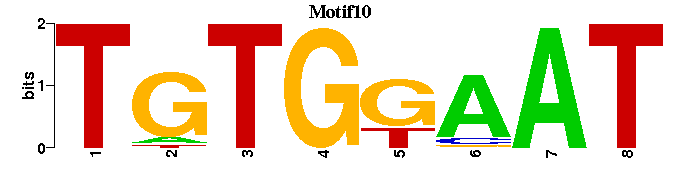 | 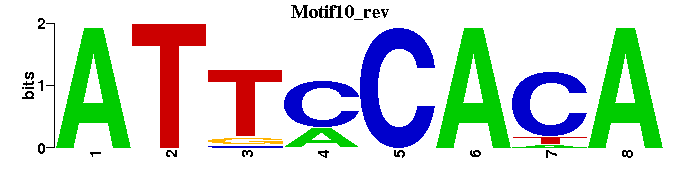 |
| Name | E value | |||
| Opaque-2_M00010 | 4.2016e-07 | YATCTACGTGGAATG | 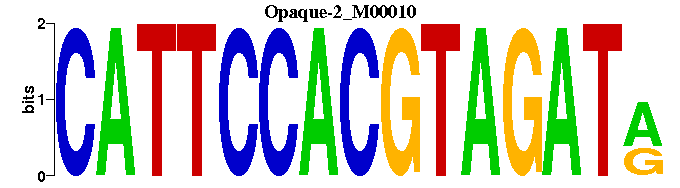 | |
| CEBP_M01087 | 2.6231e-05 | NWWNTKTGNNANYAKYYNNNNN | 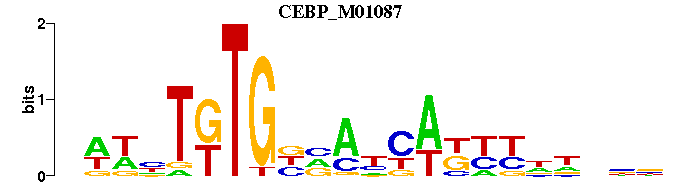 | |
| Brachyury_M00150 | 4.4115e-05 | NNNTSACACCTAGGTGTGAAATT | 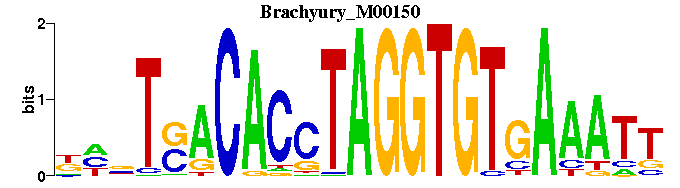 | |
| GCR1_M00046 | 6.8147e-05 | -GWGGAAGC | 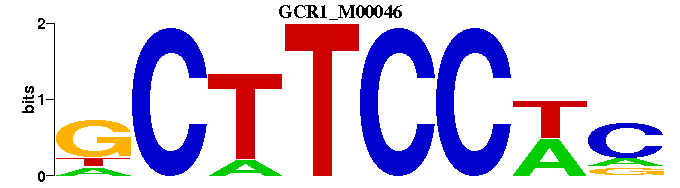 | |
| HP1_M00725 | 1.9602e-04 | CTGTTGAAWAT | 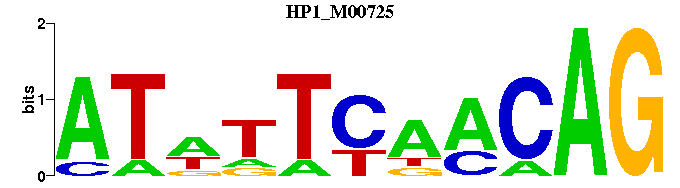 |
| Motif8 | 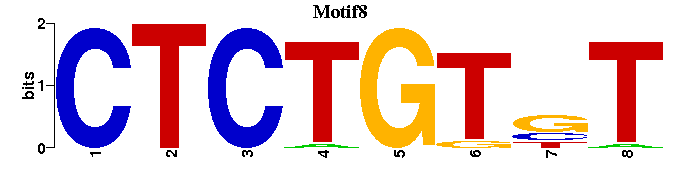 | 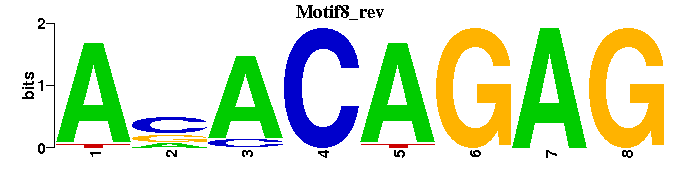 |
| Name | E value | |||
| SMAD3_M00701 | 5.7980e-06 | AGNCAGAC | 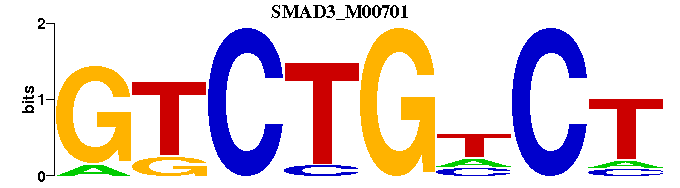 | |
| STE11_M01005 | 3.6854e-04 | TTTCTTTGTTC |  | |
| Mat1-Mc_M00275 | 4.6901e-04 | NRMNNAACAAAGANRYC |  | |
| SEF-1_M00214 | 5.4885e-04 | ACACGGATATCTGTGGTY |  | |
| SOX_M01014 | 6.8173e-04 | -TCTTTGTTANGN |  |
| Motif9 | 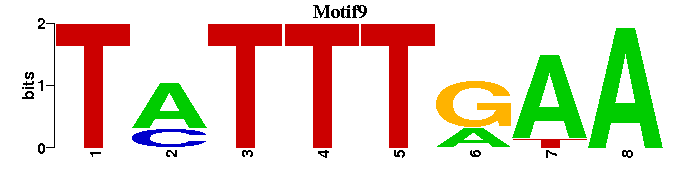 | 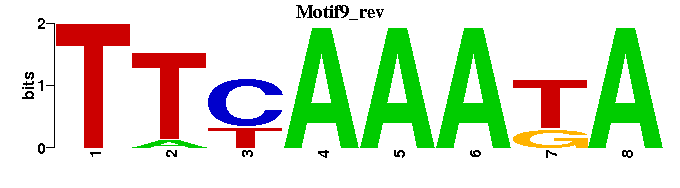 |
| Name | E value | |||
| TCF-4_M00671 | 1.5985e-06 | WTCAAAG- |  | |
| avian_M00311 | 4.6443e-05 | GCTTAWATA |  | |
| Oct-1_M00342 | 1.5921e-04 | NATTTGCAT |  | |
| Octamer_M00795 | 2.2191e-04 | NATGCAAATN | 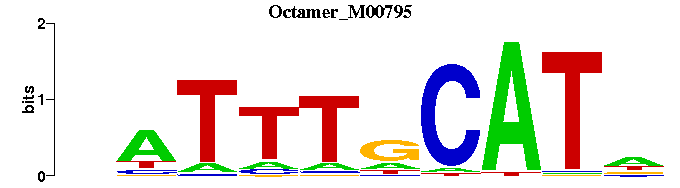 | |
| Oct-1_M00930 | 2.6329e-04 | NATGNAAATN | 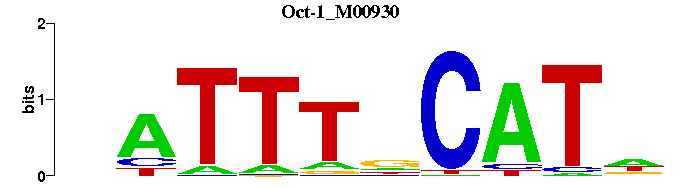 |
| Motif2 | 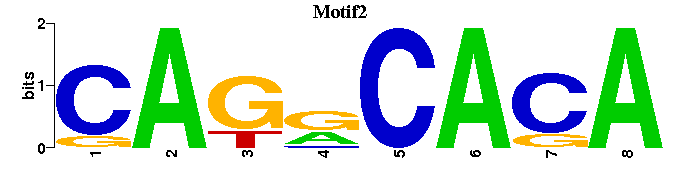 | 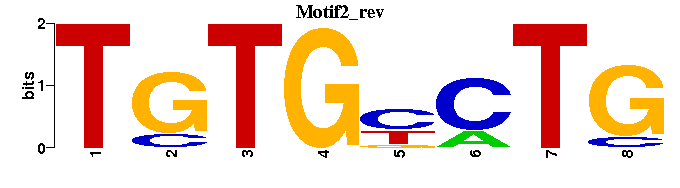 |
| Name | E value | |||
| TEIL_M00502 | 4.2676e-05 | -AGRTWCA |  | |
| SMAD3_M00701 | 2.2445e-04 | -AGNCAGAC |  | |
| Hmx3_M00433 | 4.3915e-04 | CACGCACTT |  | |
| Nrf-1_M00652 | 1.2960e-03 | GCRTGCGCR |  | |
| Zta_M00711 | 1.3054e-03 | TGWGYCANNNTN |  |
| Motif5 | 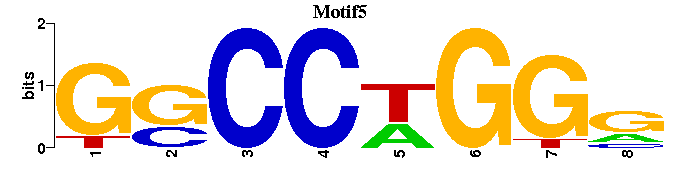 | 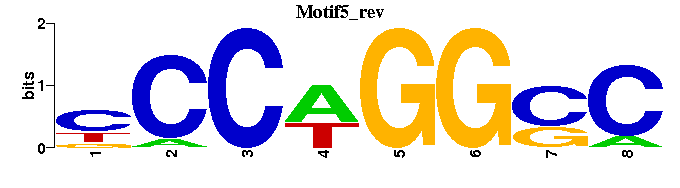 |
| Name | E value | |||
| AP-2_M00915 | 3.9977e-05 | NGCCNGNGGNNN | 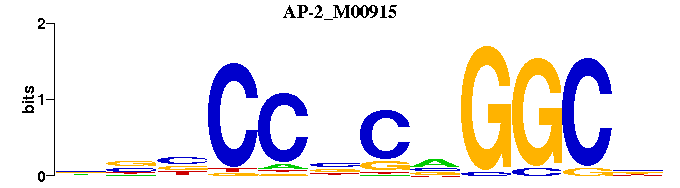 | |
| CAC-binding_M00720 | 6.0998e-04 | CCCASCCY |  | |
| EBF_M00977 | 1.2661e-03 | NCYCWRGGGA |  | |
| LRH1_M01142 | 1.6551e-03 | NTNCAAGGYYN | 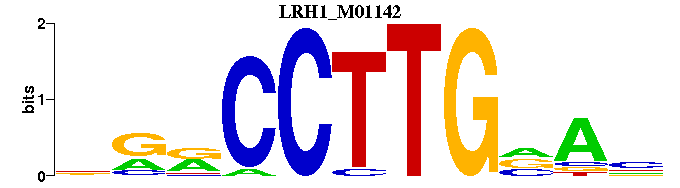 | |
| PacC_M00247 | 2.0442e-03 | NNNNNNKCYTGGCNGN |  |
| Motif4 | 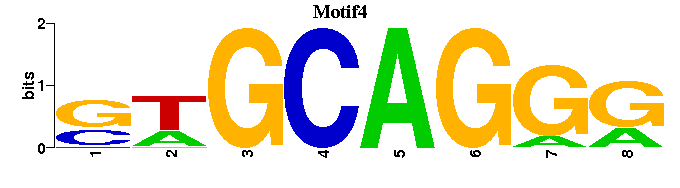 | 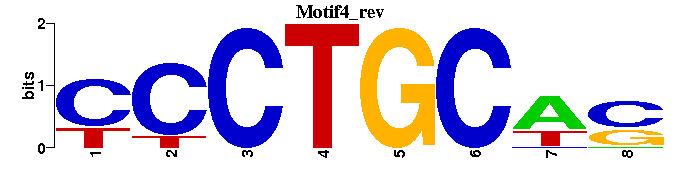 |
| Name | E value | |||
| AR_M00481 | 3.3710e-05 | AGNACANNNTGTWC | 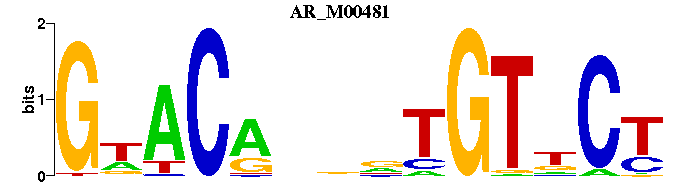 | |
| Ttk_M00009 | 1.4068e-04 | GTCCTGC-- |  | |
| Sp3_M00665 | 1.4198e-04 | CCCYSCCCAAGKS | 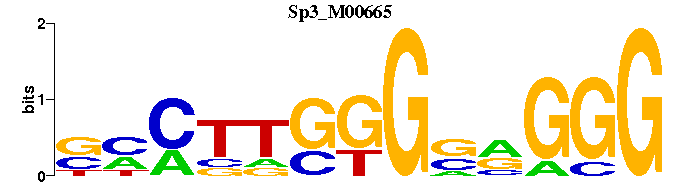 | |
| HEB_M00698 | 1.0428e-03 | CAGCWGG- |  | |
| KAISO_M01119 | 1.0885e-03 | TCCTGCNAN |  |
| Motif7 | 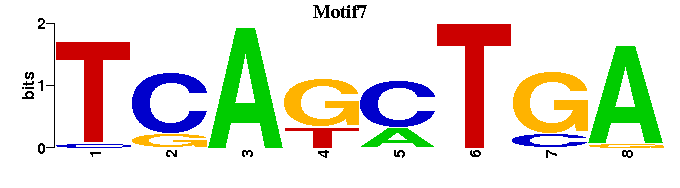 | 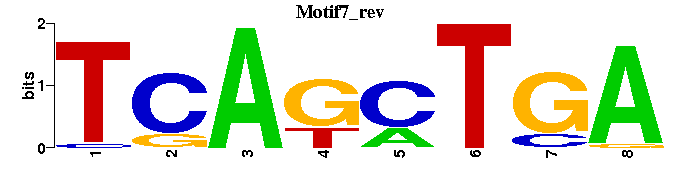 |
| Name | E value | |||
| AP-4_M00175 | 3.5844e-07 | NCAGCTGNN |  | |
| AP-4_M00176 | 9.7041e-07 | NCAGCTGNN |  | |
| HEB_M00698 | 5.5181e-06 | -CAGCWGG |  | |
| AP-4_M00927 | 7.8447e-06 | NCAGCTGY |  | |
| AP-4_M00005 | 3.7647e-05 | MNGNCNNCAGCTGNNNN |  |