 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| Motif1: | ---CCTNWCTT-- |
| Motif2: | ----TTTTCTA-- |
| Motif3: | ---GTATTGT--- |
| Motif4: | ----TGTTCTCA- |
| Motif5: | -----TTTMTAAA |
| Motif6: | -----CNTGTTT- |
| Motif7: | -----ATTCTC-- |
| Motif8: | ANAGNATT----- |
| Motif9: | --TSTNTTCT--- |
| Motif10: | ---GTAWTGT--- |
| Familial Profile: (click for matrix) | 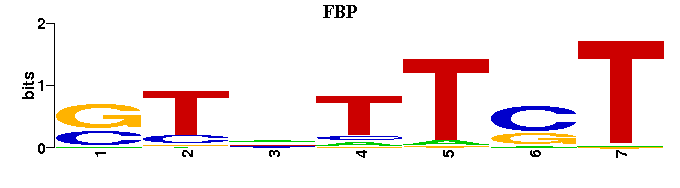 |
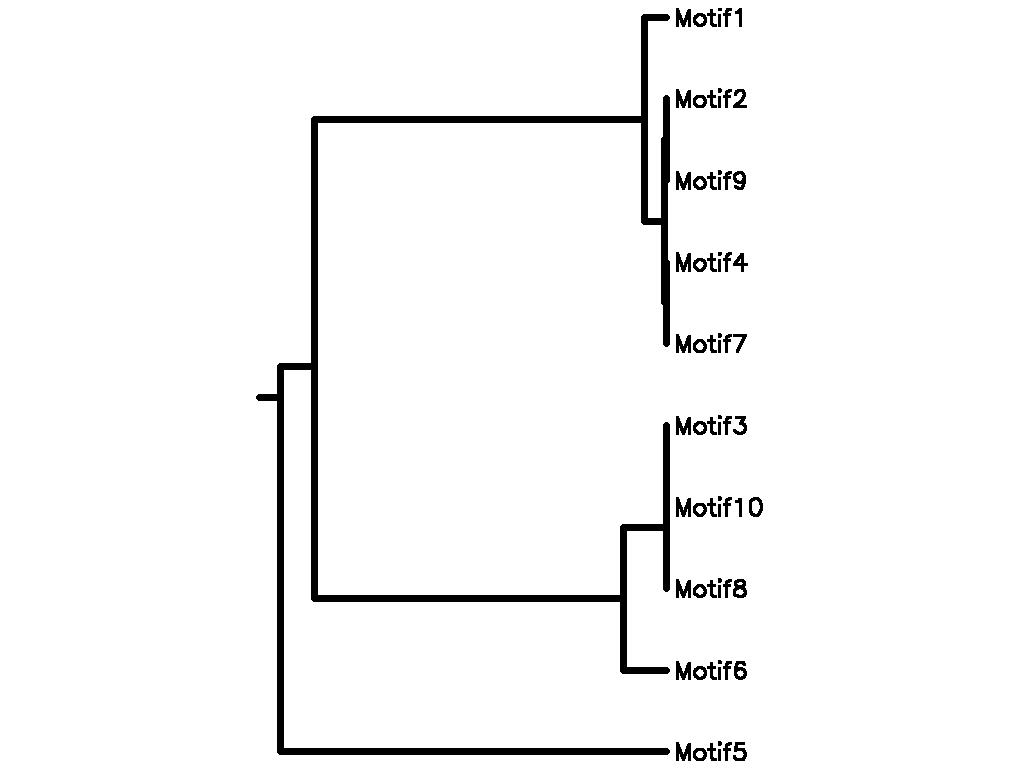
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| Motif1 | 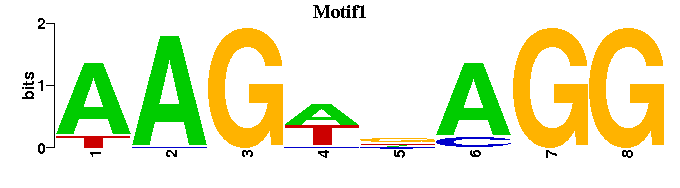 | 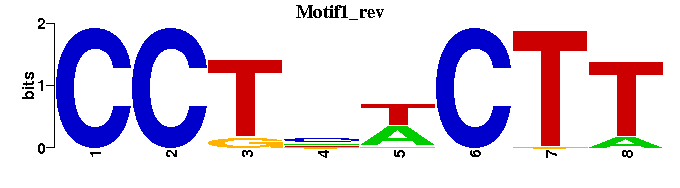 |
| Name | E value | |||
| IRF-1_M00747 | 1.4611e-07 | AANTGA-- |  | |
| Sry-beta_M00666 | 1.5886e-05 | -AGWGATGC |  | |
| IRF_M00972 | 1.3370e-04 | AAANWGAAAN | 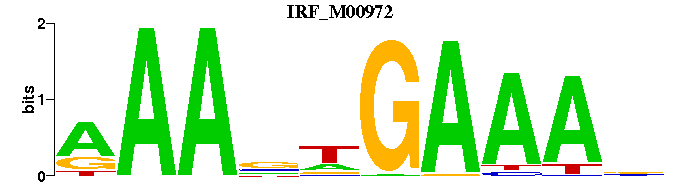 | |
| DEC_M00997 | 1.5031e-04 | NNCANGTGANNN |  | |
| TFII-I_M00706 | 2.1410e-04 | CCTMCNTC |  |
| Motif2 | 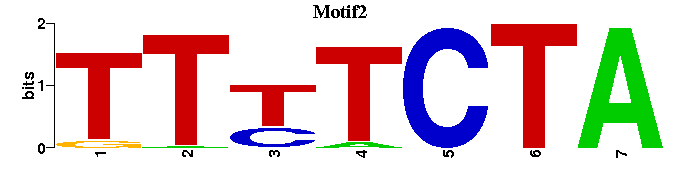 | 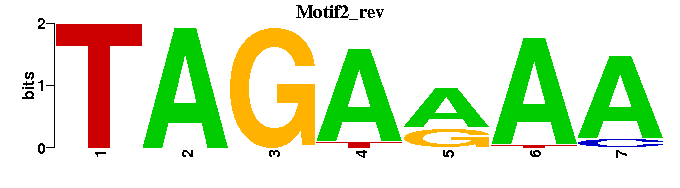 |
| Name | E value | |||
| HSF_M00166 | 2.5029e-07 | NTTCTNTTCTAGAA | 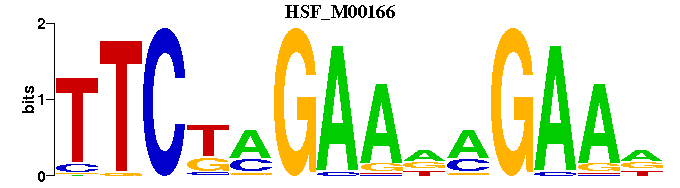 | |
| BR-C_M00092 | 7.3555e-06 | RANAAATAGNWNNNN | 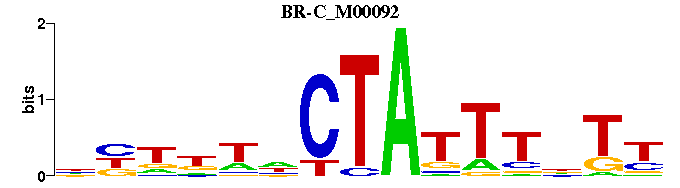 | |
| TATA_M00216 | 4.0650e-05 | NTTTTATAG |  | |
| dl_M00120 | 5.4821e-05 | NGNTTTTCYC |  | |
| HSF_M00170 | 8.3115e-05 | TTCNNGAANNGAAN | 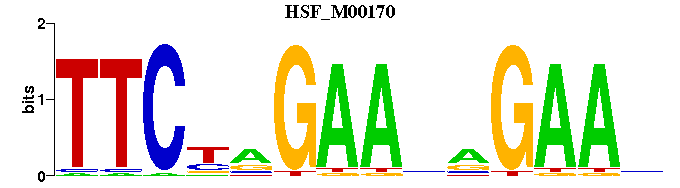 |
| Motif9 | 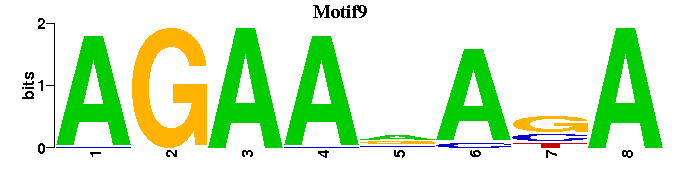 | 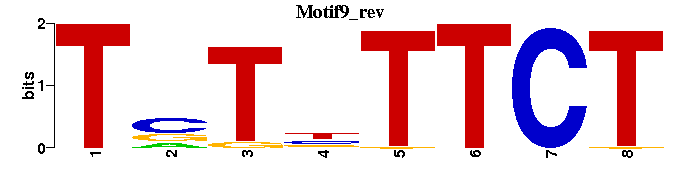 |
| Name | E value | |||
| HSF_M00163 | 6.5480e-09 | GAANAGAANAGAAN | 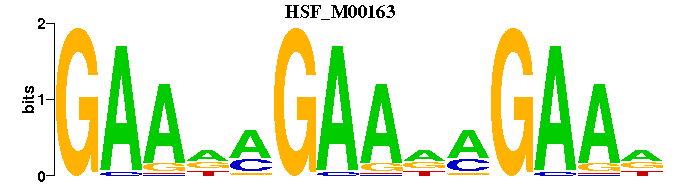 | |
| HSF_M00166 | 6.5480e-09 | TTCTAGAANAGAAN |  | |
| GATA-2_M00349 | 8.4138e-07 | TNTTATCTS |  | |
| GATA-3_M00350 | 1.8144e-06 | TNTTATCTY | 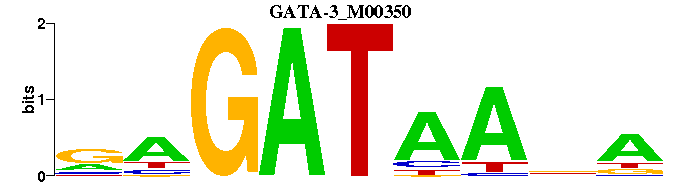 | |
| HSF_M00167 | 3.5164e-06 | NTTCNNTTCNNTTC | 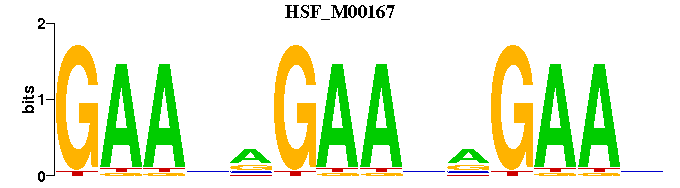 |
| Motif4 | 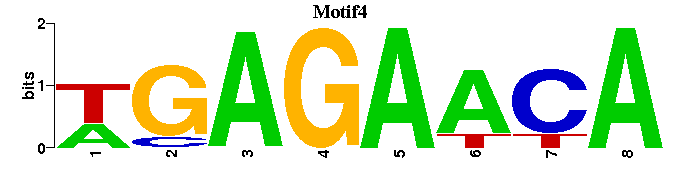 | 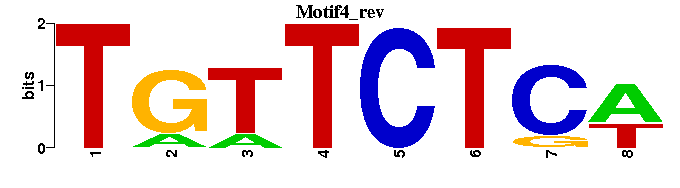 |
| Name | E value | |||
| PR,_M00960 | 9.9609e-08 | NTGTYCNNW |  | |
| Poly_M00317 | 3.7466e-07 | NAWKRRNAMACASNN | 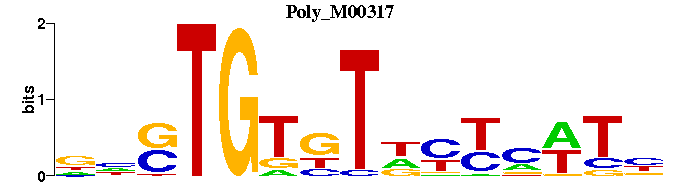 | |
| Alx-4_M00619 | 6.5339e-07 | GATTATTCTCAG |  | |
| GATA-1_M00346 | 6.6748e-07 | NNTTATCNN |  | |
| GR_M00921 | 8.4467e-07 | --AGNACAN | 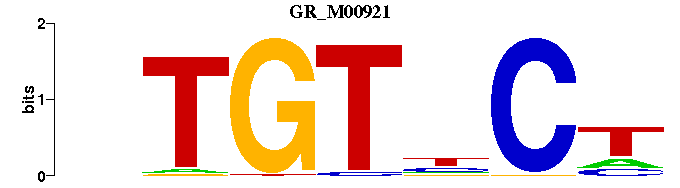 |
| Motif7 | 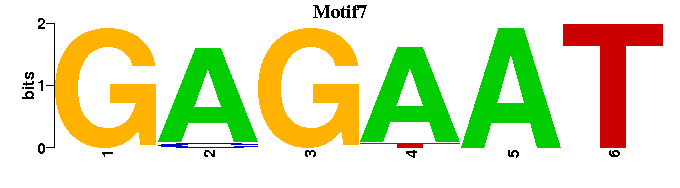 | 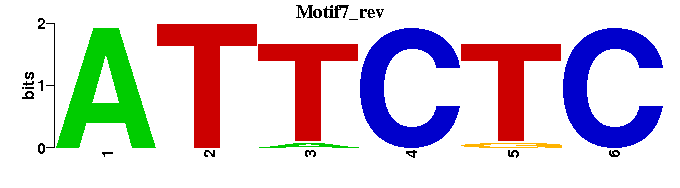 |
| Name | E value | |||
| Alx-4_M00619 | 8.7251e-08 | CTGAGAATAATC |  | |
| Ik-1_M00086 | 1.4534e-03 | NNTGGGAATNNN | 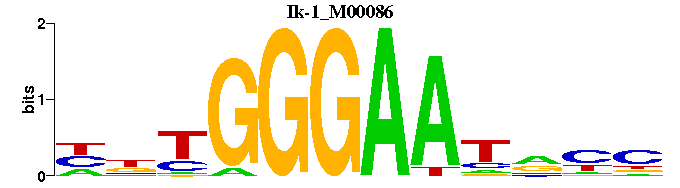 | |
| HMG_M00750 | 1.4976e-03 | AWTTTC |  | |
| dl_M00120 | 1.5382e-03 | NGNTTTTCYC |  | |
| HSF1_M01023 | 1.8404e-03 | KRAGAANNTTCYAGAA | 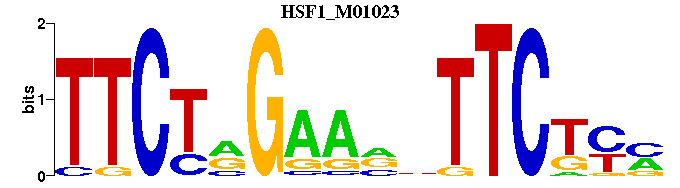 |
| Motif3 | 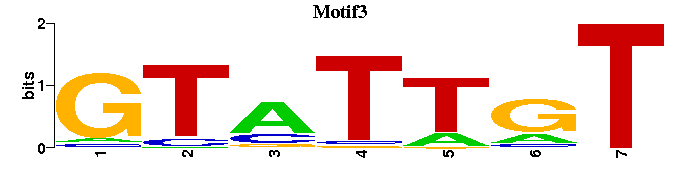 | 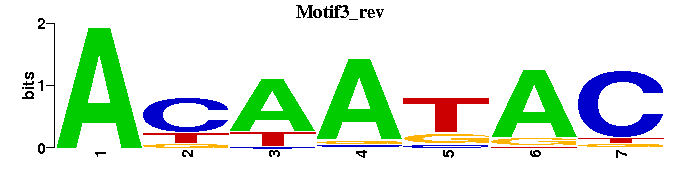 |
| Name | E value | |||
| MRF-2_M00454 | 5.2223e-09 | WSNACAATACNNM | 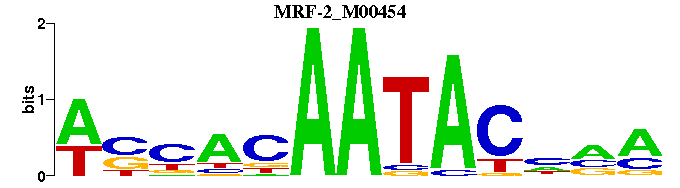 | |
| ROX1_M00728 | 1.1727e-06 | AACAATRS | 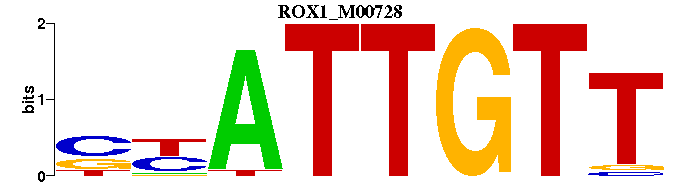 | |
| SOX17_M01016 | 1.9964e-06 | --ATTGT |  | |
| Mat1-Mc_M00276 | 1.2193e-04 | NRACAATNG |  | |
| FOXP1_M00987 | 3.0594e-04 | ATAAAAAACAACACAAATA | 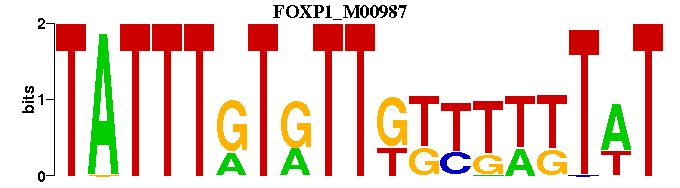 |
| Motif10 | 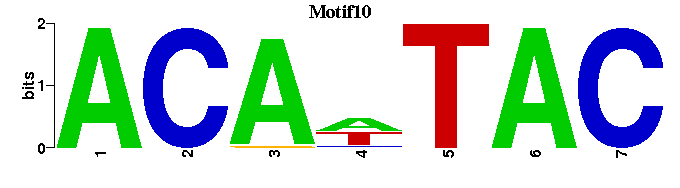 | 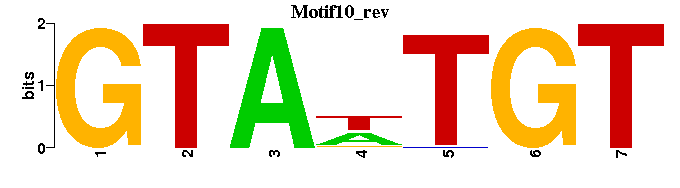 |
| Name | E value | |||
| MRF-2_M00454 | 7.2027e-08 | KNNGTATTGTNSW |  | |
| ROX1_M00728 | 1.2127e-05 | AACAATRS |  | |
| SOX17_M01016 | 1.5823e-05 | --ATTGT |  | |
| ATHB-9_M00417 | 4.4225e-04 | GNRNGTAATCATTACNNN |  | |
| Mat1-Mc_M00276 | 4.9858e-04 | NRACAATNG |  |
| Motif8 | 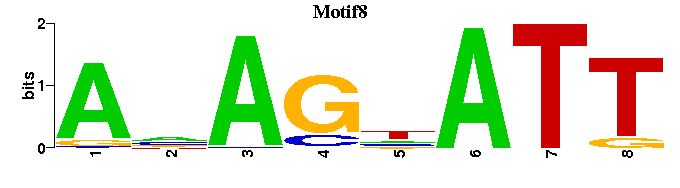 | 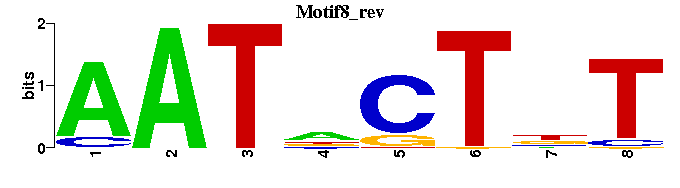 |
| Name | E value | |||
| LAC9_M00207 | 6.3834e-07 | NCGGANNRMAGWNWTCCGMKA | 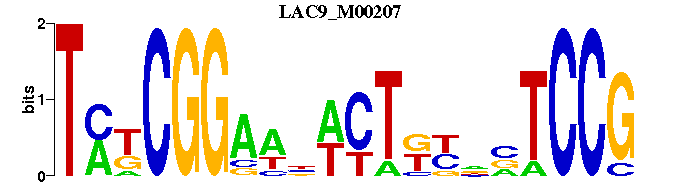 | |
| FOXJ2_M00423 | 1.0426e-05 | NMAAATATTATKR | 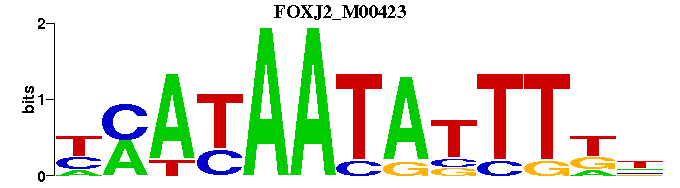 | |
| SPF1_M00702 | 5.3892e-05 | WWTACTATK |  | |
| Nkx3-1_M00451 | 7.5923e-05 | NWATACTTATW | 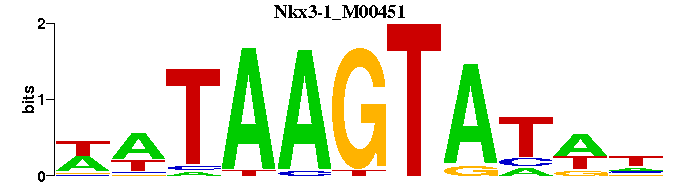 | |
| ATHB-5_M00503 | 1.4379e-04 | CAATNATT- |  |
| Motif6 |  | 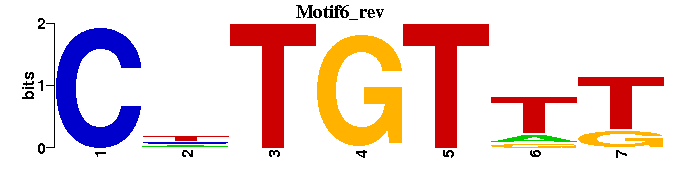 |
| Name | E value | |||
| Grainyhead-Elf-1-NTF-1_M00951 | 2.1137e-07 | AAACCRG |  | |
| HFH1_M00129 | 6.1728e-06 | AWATAAACAWT | 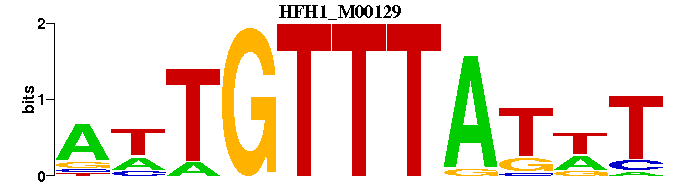 | |
| FOXO3A_M01137 | 1.0883e-05 | NNNTTGTTTAY |  | |
| FOX_M00809 | 1.2670e-05 | WAAAYAAACANT |  | |
| FOXO1_M00474 | 1.3268e-05 | NNNGTAAACAANN | 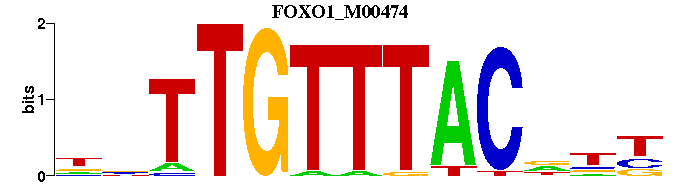 |
| Motif5 | 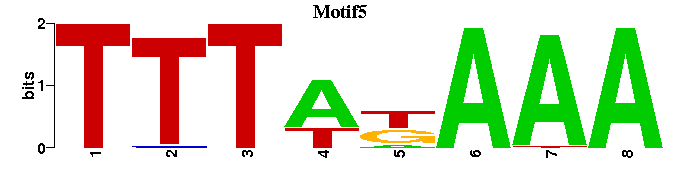 | 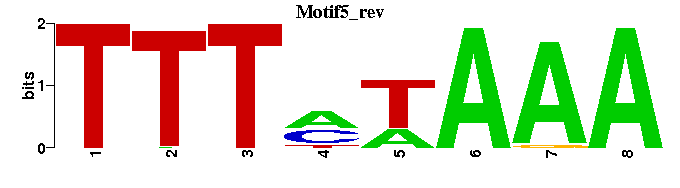 |
| Name | E value | |||
| MEF-2_M00405 | 3.4007e-06 | RGKNWTTTTTANNSM | 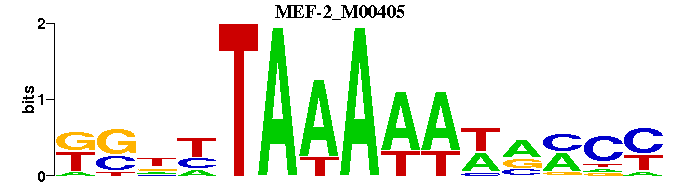 | |
| aMEF-2_M00403 | 6.7357e-06 | KKRGNTATTTTTARNCM |  | |
| C-EBPgamma_M00622 | 7.7935e-06 | WTTYTGAAATNA | 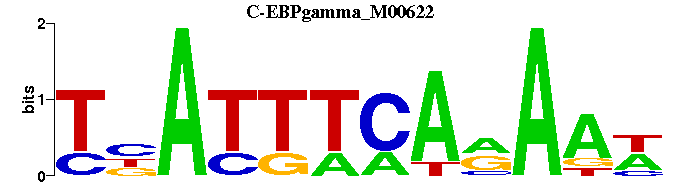 | |
| TBP_M00713 | 3.3512e-05 | NTTTATAT- | 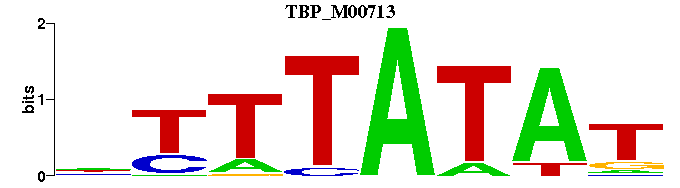 | |
| TATA_M00216 | 5.2408e-05 | NTTTTATAG- |  |