 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| Motif1: | ----WCTRNCYCT |
| Motif2: | ---AAANAAT--- |
| Motif3: | ---AAANAAT--- |
| Motif4: | ----AANATNT-- |
| Motif5: | -ANAWANAT---- |
| Motif6: | AAGAAWNAT---- |
| Motif7: | --AAWANATT--- |
| Motif8: | --WNGATAGNAA- |
| Motif9: | ---ANATATNT-- |
| Motif10: | ---ANATATNT-- |
| Familial Profile: (click for matrix) | 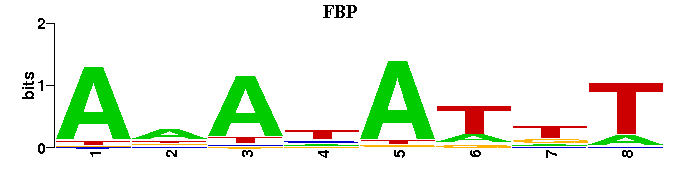 |
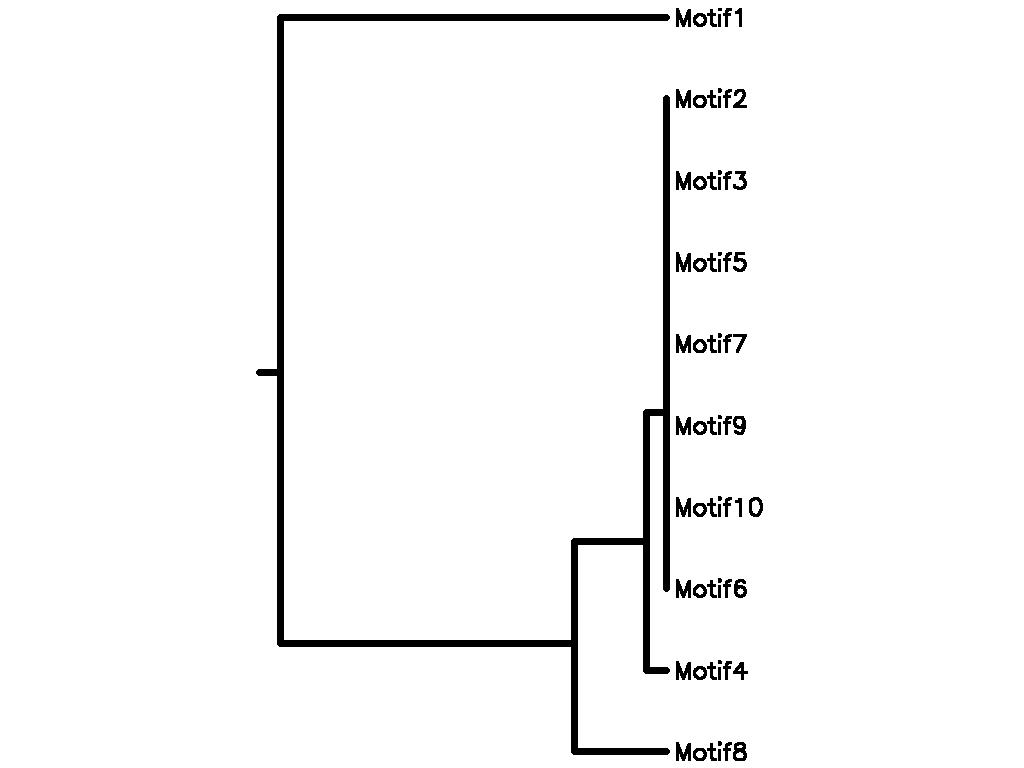
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| Motif1 | 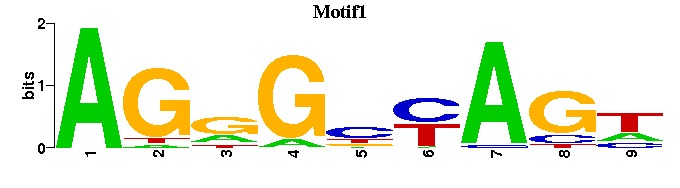 | 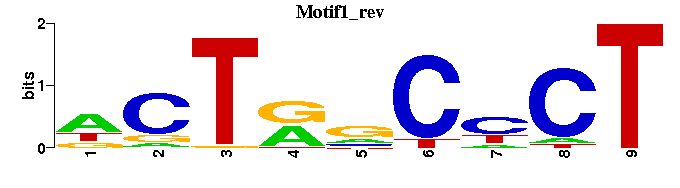 |
| Name | E value | |||
| LXR,_M00965 | 4.9828e-04 | NRGGTYANNNNAGGNC | 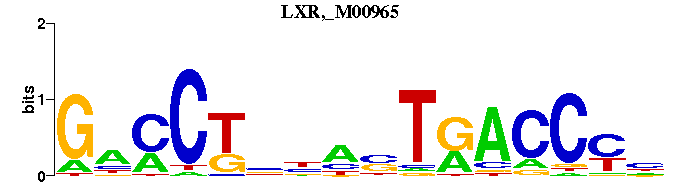 | |
| BRK_M01096 | 5.9252e-04 | -NTGGCG-- |  | |
| PXR,_M00964 | 8.8024e-04 | KNYNNTRACCC- |  | |
| NF-Y_M00209 | 1.2827e-03 | NNTARCCAATCAG | 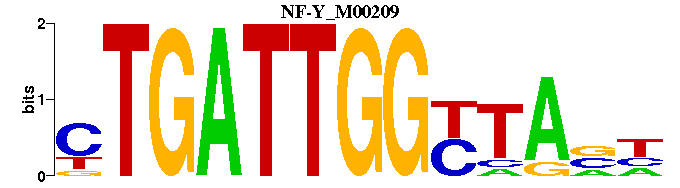 | |
| LF-A1_M00646 | 2.4147e-03 | -YWGASCC- |  |
| Motif2 | 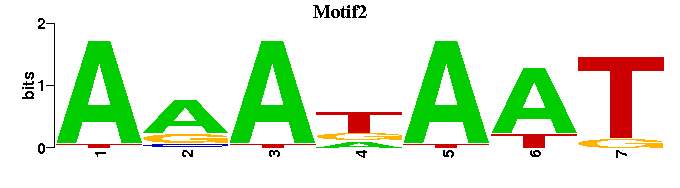 | 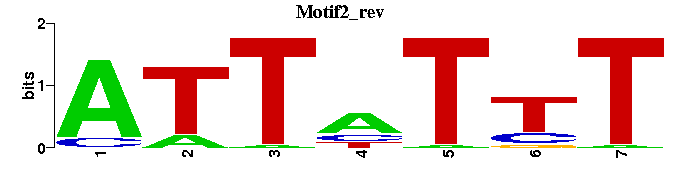 |
| Name | E value | |||
| ATHB-5_M00503 | 8.3859e-07 | AATNATTG |  | |
| XFD-1_M00267 | 1.4123e-06 | AWGTAAATAWWRY |  | |
| CDP_M00095 | 9.7541e-06 | CAATAATCGAT |  | |
| Pax-4_M00377 | 1.0186e-05 | NNNAATTWNT- |  | |
| Alx-4_M00619 | 1.2910e-05 | CTGAGAATAATC |  |
| Motif3 | 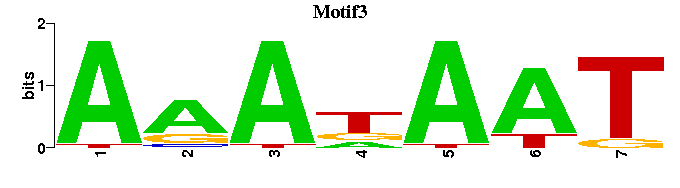 | 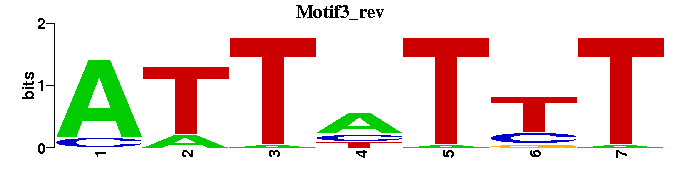 |
| Name | E value | |||
| ATHB-5_M00503 | 8.3859e-07 | AATNATTG |  | |
| XFD-1_M00267 | 1.4123e-06 | AWGTAAATAWWRY |  | |
| CDP_M00095 | 9.7541e-06 | CAATAATCGAT |  | |
| Pax-4_M00377 | 1.0186e-05 | NNNAATTWNT- |  | |
| Alx-4_M00619 | 1.2910e-05 | CTGAGAATAATC |  |
| Motif5 | 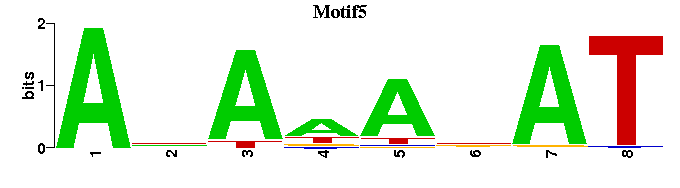 | 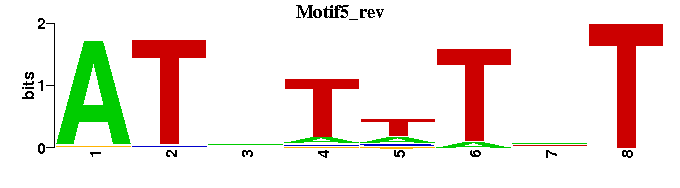 |
| Name | E value | |||
| Croc_M00266 | 2.9160e-08 | NNNNMTATTTATTNT |  | |
| RSRFC4_M00026 | 5.1956e-07 | NNKCTATAAATAGMN | 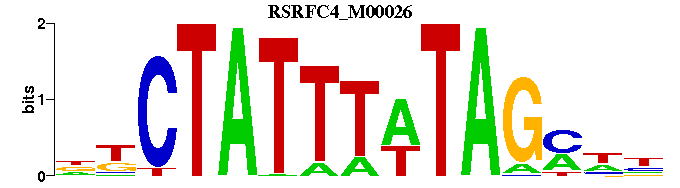 | |
| RSRFC4_M00407 | 9.9279e-07 | NNNKCTATTTWTAGMN | 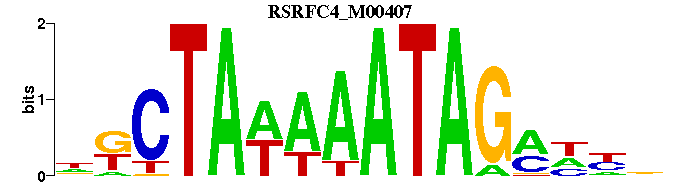 | |
| MEF-2_M00941 | 2.6014e-06 | NTAAAAATAGC |  | |
| MEF-2_M00232 | 2.7064e-06 | NNNNKCTATTTWTAGMWNNNN | 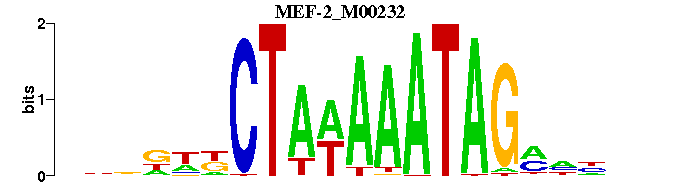 |
| Motif7 | 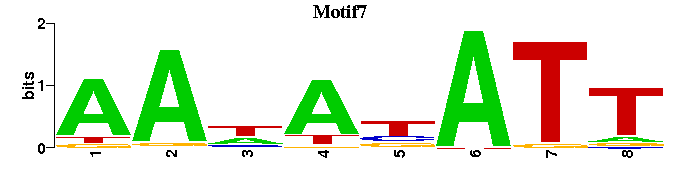 |  |
| Name | E value | |||
| FOXJ2_M00423 | 5.6533e-07 | NMAAATATTATKR | 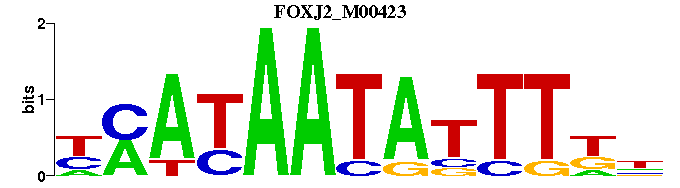 | |
| POU1F1_M00744 | 8.9834e-06 | -WWTTATNCA | 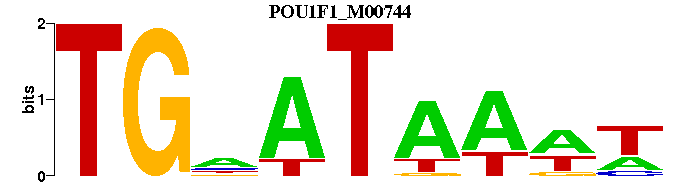 | |
| CF2-II_M00013 | 1.8538e-05 | TATATATA |  | |
| CF2-II_M00012 | 7.8772e-05 | TATATRTA |  | |
| POU3F2_M00463 | 1.2793e-04 | ATGAATWAATKCA |  |
| Motif9 | 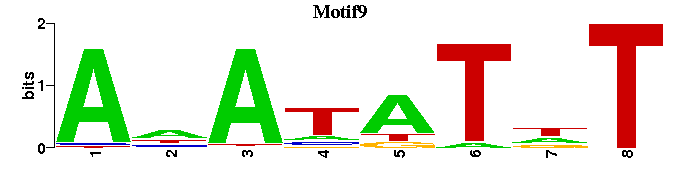 | 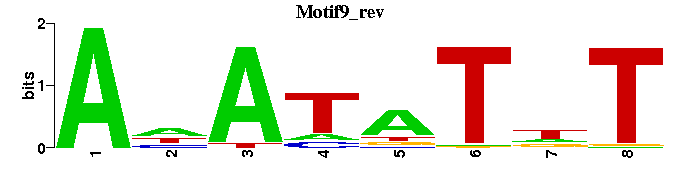 |
| Name | E value | |||
| HNF3beta_M00131 | 4.5691e-08 | GNANTRTTTRYTTW | 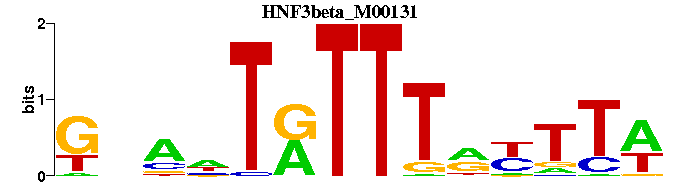 | |
| FOXJ2_M00423 | 3.2854e-06 | NMAAATATTATKR |  | |
| POU1F1_M00744 | 2.6947e-05 | WWTTATNCA |  | |
| XFD-1_M00267 | 3.7617e-05 | RYWWTATTTACWT |  | |
| ATHB-5_M00503 | 1.0234e-04 | AATNATTG |  |
| Motif10 | 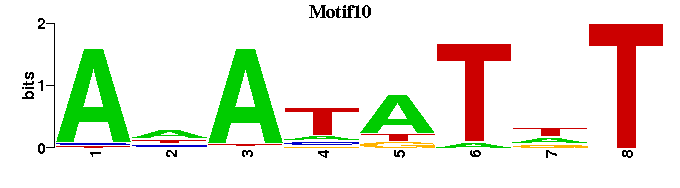 | 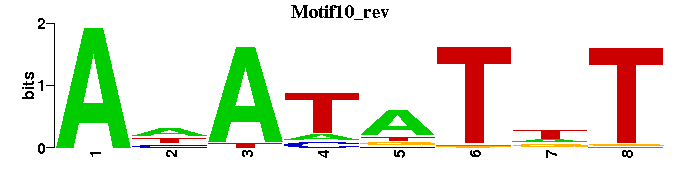 |
| Name | E value | |||
| HNF3beta_M00131 | 4.5691e-08 | GNANTRTTTRYTTW |  | |
| FOXJ2_M00423 | 3.2854e-06 | NMAAATATTATKR |  | |
| POU1F1_M00744 | 2.6947e-05 | WWTTATNCA |  | |
| XFD-1_M00267 | 3.7617e-05 | RYWWTATTTACWT |  | |
| ATHB-5_M00503 | 1.0234e-04 | AATNATTG |  |
| Motif6 | 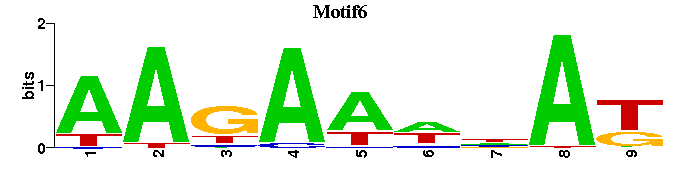 | 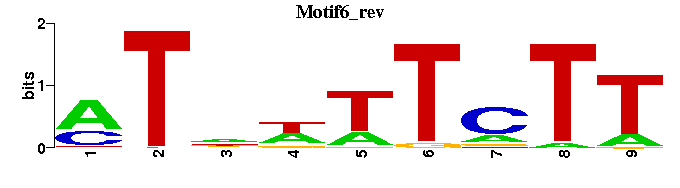 |
| Name | E value | |||
| Alx-4_M00619 | 4.8131e-06 | GATTATTCTCAG |  | |
| Croc_M00266 | 1.0535e-05 | NNNNMTATTTATTNT |  | |
| Rim101p_M01030 | 8.1075e-05 | ---TTNCTTG | 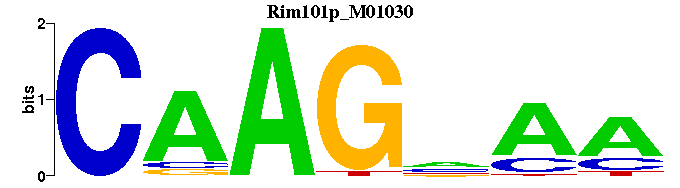 | |
| HSF_M00164 | 9.3824e-05 | AGAANNTTCTNTTC | 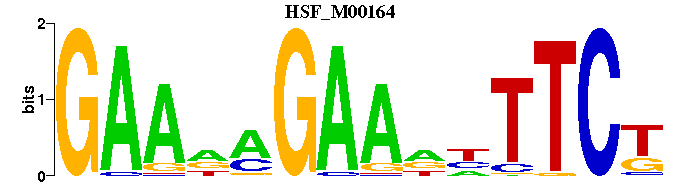 | |
| POU1F1_M00744 | 2.1434e-04 | TGNATAAWW-- |  |
| Motif4 | 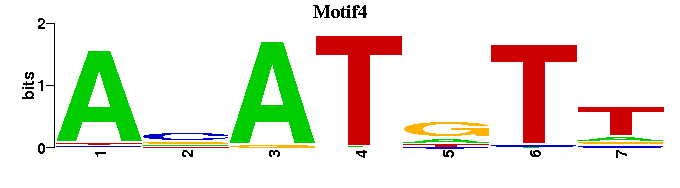 | 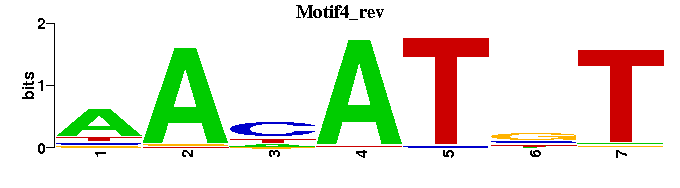 |
| Name | E value | |||
| AR_M00447 | 1.9348e-06 | GWACNTNWTGTTCT | 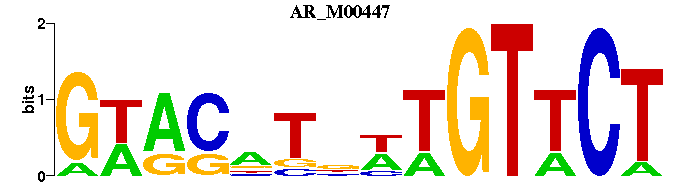 | |
| RP58_M00532 | 1.9770e-05 | TCCAGATGTTN | 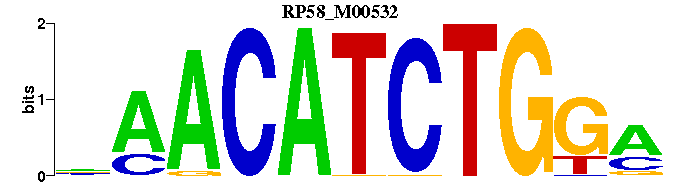 | |
| TFE_M01029 | 8.1241e-05 | NCACATG-- |  | |
| p53_M00272 | 1.3045e-04 | RGRCAWGYC |  | |
| MATalpha2_M00031 | 1.3146e-04 | -CRTGTNNWN | 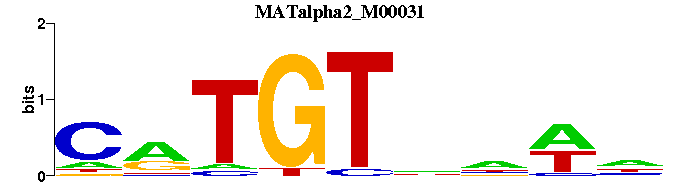 |
| Motif8 | 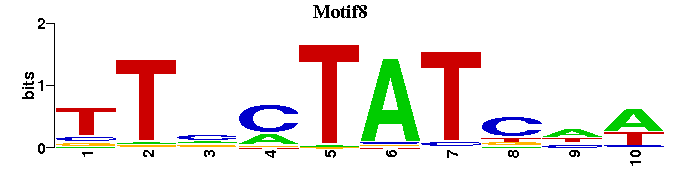 | 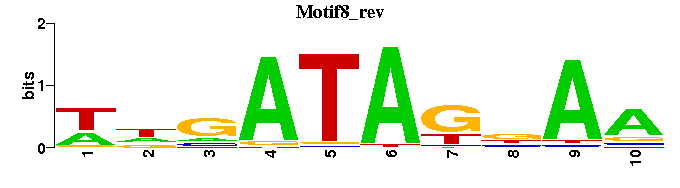 |
| Name | E value | |||
| GATA-2_M00349 | 4.0554e-04 | -TNTTATCTS |  | |
| Staf_M00264 | 7.6002e-04 | NNGCNNKGNANNNTGGGANN |  | |
| GATA-3_M00350 | 8.2084e-04 | RAGATAANA- | 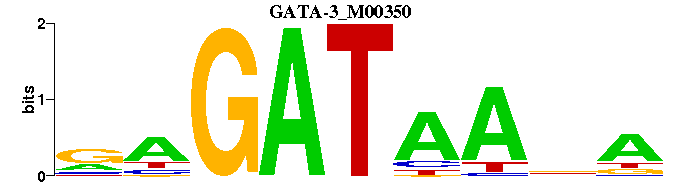 | |
| Evi-1_M00081 | 8.5604e-04 | TATCTTNTCNTATC |  | |
| Pbx-1_M00096 | 1.0884e-03 | TTGATTRN-- | 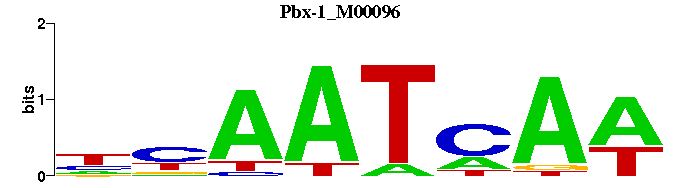 |